bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2277_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
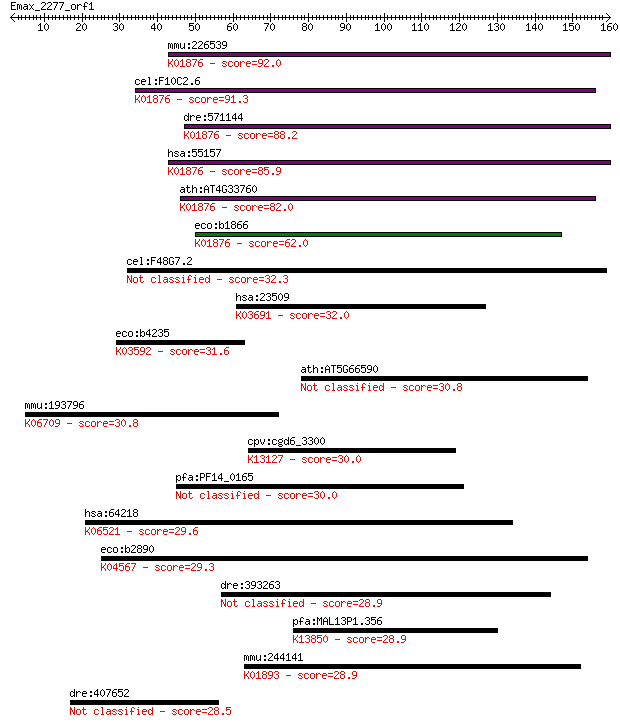
mmu:226539 Dars2, 5830468K18Rik, MGC99963; aspartyl-tRNA synth... 92.0 7e-19
cel:F10C2.6 drs-2; aspartyl(D) tRNA Synthetase family member (... 91.3 1e-18
dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase 2,... 88.2 1e-17
hsa:55157 DARS2, ASPRS, FLJ10514, LBSL, MT-ASPRS, RP3-383J4.2;... 85.9 5e-17
ath:AT4G33760 tRNA synthetase class II (D, K and N) family pro... 82.0 8e-16
eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase... 62.0 8e-10
cel:F48G7.2 hypothetical protein 32.3 0.63
hsa:23509 POFUT1, FUT12, KIAA0180, MGC2482, O-FUT, O-Fuc-T, O-... 32.0 0.97
eco:b4235 pmbA, ECK4230, JW4194, mcb, tldE; predicted peptidas... 31.6 1.2
ath:AT5G66590 allergen V5/Tpx-1-related family protein 30.8 2.0
mmu:193796 Kdm4b, 4732474L06Rik, Jmjd2b, MGC6349, mKIAA0876; l... 30.8 2.1
cpv:cgd6_3300 Yir323cp/Cwc24 p family; CCCH+ringfinger domains... 30.0 3.1
pfa:PF14_0165 conserved Plasmodium protein, unknown function 30.0 3.7
hsa:64218 SEMA4A, CORD10, FLJ12287, RP35, SEMAB, SEMB; sema do... 29.6 3.9
eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synth... 29.3 5.1
dre:393263 MGC56488, ddrgk1, zgc:77356; zgc:56488 28.9 6.7
pfa:MAL13P1.356 VAR; erythrocyte membrane protein 1, PfEMP1; K... 28.9 7.2
mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthet... 28.9 8.5
dre:407652 MGC123116, wu:fj19d07; zgc:123116 28.5 8.5
> mmu:226539 Dars2, 5830468K18Rik, MGC99963; aspartyl-tRNA synthetase
2 (mitochondrial) (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=653
Score = 92.0 bits (227), Expect = 7e-19, Method: Composition-based stats.
Identities = 47/121 (38%), Positives = 70/121 (57%), Gaps = 5/121 (4%)
Query 43 DVNRFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDSYGSVQLFVKSSELQEI 102
+ + F R++ CG++ S +G+EVTLCGWI+Y R FL LRD +G VQ+ + E
Sbjct 42 EFSSFVARTNTCGELRSSHLGQEVTLCGWIQYRRQNTFLVLRDCHGLVQILIPQDESAAS 101
Query 103 VRNL----SLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNMCKANLPFLNRD 158
VR + +ESV++ G V RP N K+ G +E+ V+ +LN CK LPF +D
Sbjct 102 VRRILCEAPVESVVRVSGTVISRPPGQENPKMPTGEIEIKVKTAELLNACK-KLPFEIKD 160
Query 159 Y 159
+
Sbjct 161 F 161
> cel:F10C2.6 drs-2; aspartyl(D) tRNA Synthetase family member
(drs-2); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=593
Score = 91.3 bits (225), Expect = 1e-18, Method: Composition-based stats.
Identities = 49/128 (38%), Positives = 80/128 (62%), Gaps = 7/128 (5%)
Query 34 TANSVKAEC-----DVNRFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDSYG 88
+A +V A C + +T R+H C ++ S E+V++ GW+ + R +F LRD+YG
Sbjct 3 SARNVTAICCRRILSTSSYTVRTHICDELSTSNKNEKVSVMGWLSHKRMDRFFVLRDAYG 62
Query 89 SVQLFVK-SSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNM 147
SVQ + SS+LQ +++++ ESV++ G V R D + NSK+ G++E+ VEEL+VLN
Sbjct 63 SVQAKISASSKLQSLLKDIPYESVVRVDGIVVDRGD-NRNSKMKTGDIEIDVEELTVLNK 121
Query 148 CKANLPFL 155
+N+P L
Sbjct 122 ATSNIPML 129
> dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase
2, mitochondrial; K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=660
Score = 88.2 bits (217), Expect = 1e-17, Method: Composition-based stats.
Identities = 47/117 (40%), Positives = 71/117 (60%), Gaps = 5/117 (4%)
Query 47 FTYRSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDSYGSVQLFV----KSSELQEI 102
F+ RSH CG++ S + +EVTLCGW++Y R F+ LRD G VQ+ + SEL++
Sbjct 68 FSQRSHTCGELCSSHIDKEVTLCGWVQYLRQDLFVILRDFSGLVQILIPQDESKSELKKS 127
Query 103 VRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNMCKANLPFLNRDY 159
+ L++ESVI KG+V RP+ N + G +EV E + VLN + LPF +++
Sbjct 128 LLALTVESVIMVKGRVIRRPEGQENKNISTGEIEVCAESIEVLNTSR-KLPFEIKEF 183
> hsa:55157 DARS2, ASPRS, FLJ10514, LBSL, MT-ASPRS, RP3-383J4.2;
aspartyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=645
Score = 85.9 bits (211), Expect = 5e-17, Method: Composition-based stats.
Identities = 45/121 (37%), Positives = 71/121 (58%), Gaps = 5/121 (4%)
Query 43 DVNRFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDSYGSVQLFVKSSE---- 98
+ + F R++ CG++ S +G+EVTLCGWI+Y R FL LRD G VQ+ + E
Sbjct 43 EFSSFVVRTNTCGELRSSHLGQEVTLCGWIQYRRQNTFLVLRDFDGLVQVIIPQDESAAS 102
Query 99 LQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNMCKANLPFLNRD 158
+++I+ +ESV++ G V RP N K+ G +E+ V+ +LN CK LPF ++
Sbjct 103 VKKILCEAPVESVVQVSGTVISRPAGQENPKMPTGEIEIKVKTAELLNACK-KLPFEIKN 161
Query 159 Y 159
+
Sbjct 162 F 162
> ath:AT4G33760 tRNA synthetase class II (D, K and N) family protein
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=664
Score = 82.0 bits (201), Expect = 8e-16, Method: Composition-based stats.
Identities = 44/116 (37%), Positives = 64/116 (55%), Gaps = 6/116 (5%)
Query 46 RFTYRSHCCGQVDDSLVGEEVTLCGWIEYHRF---CKFLTLRDSYGSVQLFVKSSELQE- 101
R+ R+ CG++ + VG+ V LCGW+ HR FL LRD G VQ+ E E
Sbjct 71 RWVSRTELCGELSVNDVGKRVHLCGWVALHRVHGGLTFLNLRDHTGIVQVRTLPDEFPEA 130
Query 102 --IVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLNMCKANLPFL 155
++ ++ LE V+ +G V RP++ VN K+ G VEV E + +LN + LPFL
Sbjct 131 HGLINDMRLEYVVLVEGTVRSRPNESVNKKMKTGFVEVVAEHVEILNPVRTKLPFL 186
> eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=590
Score = 62.0 bits (149), Expect = 8e-10, Method: Composition-based stats.
Identities = 34/102 (33%), Positives = 55/102 (53%), Gaps = 5/102 (4%)
Query 50 RSHCCGQVDDSLVGEEVTLCGWIEYHRFCK---FLTLRDSYGSVQLFV--KSSELQEIVR 104
R+ CGQ+ S VG++VTLCGW+ R F+ +RD G VQ+F ++ ++
Sbjct 2 RTEYCGQLRLSHVGQQVTLCGWVNRRRDLGSLIFIDMRDREGIVQVFFDPDRADALKLAS 61
Query 105 NLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLN 146
L E I+ G V R +K++N + G +EV L+++N
Sbjct 62 ELRNEFCIQVTGTVRARDEKNINRDMATGEIEVLASSLTIIN 103
> cel:F48G7.2 hypothetical protein
Length=149
Score = 32.3 bits (72), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 62/132 (46%), Gaps = 18/132 (13%)
Query 32 QHTANSVKAECDVNRFTYRSHCCGQVDDSLVGEEVTLCG-WIEYHRF---CKFLTLRDSY 87
Q ++N + ECDV+ RS C Q+ EE+ L +E R K+L +
Sbjct 5 QTSSNDICQECDVSEIGQRSSNC-QIASIRANEEILLRAIQLEDQRINDSKKYLFSTHTR 63
Query 88 GSVQLFVKSSE-LQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNGNVEVSVEELSVLN 146
+Q F K+ E L +++RNL+ E IK + + P +V +V+G VE
Sbjct 64 EVIQKFHKTFEPLDDVLRNLN-EIYIKCIPEAGFFP--EVKKGVVDGFVE---------K 111
Query 147 MCKANLPFLNRD 158
+ ANL F NR+
Sbjct 112 IADANLSFKNRN 123
> hsa:23509 POFUT1, FUT12, KIAA0180, MGC2482, O-FUT, O-Fuc-T,
O-FucT-1; protein O-fucosyltransferase 1 (EC:2.4.1.221); K03691
peptide-O-fucosyltransferase [EC:2.4.1.221]
Length=388
Score = 32.0 bits (71), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 31/67 (46%), Gaps = 4/67 (5%)
Query 61 LVGEEVTLCGWIEY-HRFCKFLTLRDSYGSVQLFVKSSELQEIVRNLSLESVIKDKGKVN 119
L+ + + WIEY H F L SY Q + K LQ R +SLE ++ +
Sbjct 60 LLNRTLAVPPWIEYQHHKPPFTNLHVSY---QKYFKLEPLQAYHRVISLEDFMEKLAPTH 116
Query 120 YRPDKDV 126
+ P+K V
Sbjct 117 WPPEKRV 123
> eco:b4235 pmbA, ECK4230, JW4194, mcb, tldE; predicted peptidase
required for the maturation and secretion of the antibiotic
peptide MccB17; K03592 PmbA protein
Length=450
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 29 LNPQHTANSVKAECDVNRFTYRSHCCGQVDDSLV 62
L+PQ A +V+A D+ R+T C G D L+
Sbjct 84 LSPQAIARTVQAALDIARYTSPDPCAGVADKELL 117
> ath:AT5G66590 allergen V5/Tpx-1-related family protein
Length=185
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 43/91 (47%), Gaps = 17/91 (18%)
Query 78 CKFLTLRDS-YGSVQLFVKSSELQEIVRNLSLESVIKDKGKVNYRPD-----------KD 125
C+F +L YG+ QL+ K L + +L++E+ +K+K NY+ D K
Sbjct 88 CEFASLNPGKYGANQLWAKG--LVAVTPSLAVETWVKEKPFYNYKSDTCAANHTCGVYKQ 145
Query 126 V---NSKLVNGNVEVSVEELSVLNMCKANLP 153
V NSK + +E +VL +C N P
Sbjct 146 VVWRNSKELGCAQATCTKESTVLTICFYNPP 176
> mmu:193796 Kdm4b, 4732474L06Rik, Jmjd2b, MGC6349, mKIAA0876;
lysine (K)-specific demethylase 4B; K06709 jumonji domain-containing
protein 2 [EC:1.14.11.-]
Length=1086
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 5/72 (6%)
Query 5 SSVNPASQALSRNHSL-PQCCFLSTLNPQHT--ANSVKAECDVNRFTYRSHCCGQVDDSL 61
S+V P S++ R L P+ CF S+ ANS E + +HCC QV S
Sbjct 700 SAVQPPSKSGQRTRPLIPEMCFTSSGENTEPLPANSYVGEDGTSPLISCAHCCLQVHASC 759
Query 62 VG--EEVTLCGW 71
G E+ GW
Sbjct 760 YGVRPELAKEGW 771
> cpv:cgd6_3300 Yir323cp/Cwc24 p family; CCCH+ringfinger domains
; K13127 RING finger protein 113A
Length=311
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query 64 EEVTLCGWIEYHRFCKFLTLRDSYGSVQLFVKSSELQEIVRNLSLESVIKDKGKV 118
+E CG+ + CKFL R + S K E+++ + L +ES+ KDK +
Sbjct 161 KETGYCGFGDT---CKFLHDRSDFKSGWKLDKEWEIEQKKKRLKIESISKDKSSI 212
> pfa:PF14_0165 conserved Plasmodium protein, unknown function
Length=3026
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 35/77 (45%), Gaps = 12/77 (15%)
Query 45 NRFTY-RSHCCGQVDDSLVGEEVTLCGWIEYHRFCKFLTLRDSYGSVQLFVKSSELQEIV 103
N+FT + C VD+ + IEY FCK+ Y S S++ E++
Sbjct 1537 NKFTQVKYENCNNVDERKITPTFETPSGIEYPSFCKY-----DYNST-----SNQKIEVI 1586
Query 104 RNLSLESVIKDKGKVNY 120
+ +E V KDK K NY
Sbjct 1587 NKVCMEEV-KDKKKENY 1602
> hsa:64218 SEMA4A, CORD10, FLJ12287, RP35, SEMAB, SEMB; sema
domain, immunoglobulin domain (Ig), transmembrane domain (TM)
and short cytoplasmic domain, (semaphorin) 4A; K06521 semaphorin
4
Length=761
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 27/116 (23%), Positives = 47/116 (40%), Gaps = 19/116 (16%)
Query 21 PQCCFLSTLNPQHTANSVKAECDVNRFTYRSHCCGQVDDSLVGEEVT---LCGWIEYHRF 77
P L + P ++ K+EC + + + C + LV VT CG +
Sbjct 92 PGVPRLKNMIPWPASDRKKSECAFKKKSNETQCFNFIR-VLVSYNVTHLYTCGTFAFSPA 150
Query 78 CKFLTLRDSYGSVQLFVKSSELQEIVRNLSLESVIKDKGKVNYRPDKDVNSKLVNG 133
C F+ L+DSY + +S + V++ KG+ + P + LV+G
Sbjct 151 CTFIELQDSY---------------LLPISEDKVMEGKGQSPFDPAHKHTAVLVDG 191
> eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synthetase,
constitutive (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=505
Score = 29.3 bits (64), Expect = 5.1, Method: Composition-based stats.
Identities = 35/139 (25%), Positives = 56/139 (40%), Gaps = 30/139 (21%)
Query 25 FLSTLNPQHTANSVKAECDVNRFTYRSHCCGQVDDSL--VGEEVTLCGWIEYHRF---CK 79
F + HT++ + AE D G+ ++ L + EV + G + R
Sbjct 36 FPNDFRRDHTSDQLHAEFD-----------GKENEELEALNIEVAVAGRMMTRRIMGKAS 84
Query 80 FLTLRDSYGSVQLFVKSSELQEIVRN-----LSLESVIKDKGKVNYRPDKDVNSKLVNGN 134
F+TL+D G +QL+V +L E V N L ++ KGK+ K G
Sbjct 85 FVTLQDVGGRIQLYVARDDLPEGVYNEQFKKWDLGDILGAKGKL---------FKTKTGE 135
Query 135 VEVSVEELSVLNMCKANLP 153
+ + EL +L LP
Sbjct 136 LSIHCTELRLLTKALRPLP 154
> dre:393263 MGC56488, ddrgk1, zgc:77356; zgc:56488
Length=300
Score = 28.9 bits (63), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 11/98 (11%)
Query 57 VDDSLVGEEVT-------LCGWIEYHRFCKFLTLRDSYGSVQLFVKSS--ELQEIVRNLS 107
++D EE+T L +I+Y + K + L D L + + LQ+++ + S
Sbjct 186 IEDQGEAEELTEHESQSLLQEFIQYVQKSKVVLLEDLASQFGLRTQDAIARLQDLIADGS 245
Query 108 LESVIKDKGKVNYRPDKDVN--SKLVNGNVEVSVEELS 143
L VI D+GK + +++N ++ + VS+ EL+
Sbjct 246 LTGVIDDRGKFIFITPEELNAVAQFIKQRGRVSISELA 283
> pfa:MAL13P1.356 VAR; erythrocyte membrane protein 1, PfEMP1;
K13850 erythrocyte membrane protein 1
Length=2223
Score = 28.9 bits (63), Expect = 7.2, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Query 76 RFCKFLTLRDSYGSVQLFVKSSELQEIVRNLS-LESVIKDKGKVNYRPDKDVNSK 129
+F K L +++YG+V F+K + + ++ E +I K +YR DK++N+K
Sbjct 448 KFYKKLKEKNNYGTVDDFLKLLNKENECKGINEQEEIIDFSNKCDYRFDKNINNK 502
> mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthetase
2 (mitochondrial)(putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 28.9 bits (63), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 46/94 (48%), Gaps = 15/94 (15%)
Query 63 GEEVTLCGWIEYHRFCK---FLTLRD--SYGSVQLFVKSSELQEIVRNLSLESVIKDKGK 117
GE VT+ GWI R K FL + D S S+Q+ SS R L+ S ++ +G+
Sbjct 41 GECVTVQGWIRSVRSQKEVLFLHVNDGSSLESLQIVADSSFDS---RELTFGSSVQVQGQ 97
Query 118 VNYRPDKDVNSKLVNGNVEVSVEELSVLNMCKAN 151
+ V S+ NVE+ E++ V+ C+A
Sbjct 98 L-------VKSQSKRQNVELKAEKIEVIGDCEAK 124
> dre:407652 MGC123116, wu:fj19d07; zgc:123116
Length=752
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query 17 NHSLPQCCFLSTLNPQHTANSVK-AECDVNRFTYRSHCCG 55
NHS + C P H A+SVK ++C + Y +HC G
Sbjct 345 NHSDARFCDWCGAKPGHKASSVKCSQCGASSHPYANHCGG 384
Lambda K H
0.318 0.131 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40