bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2250_orf2
Length=93
Score E
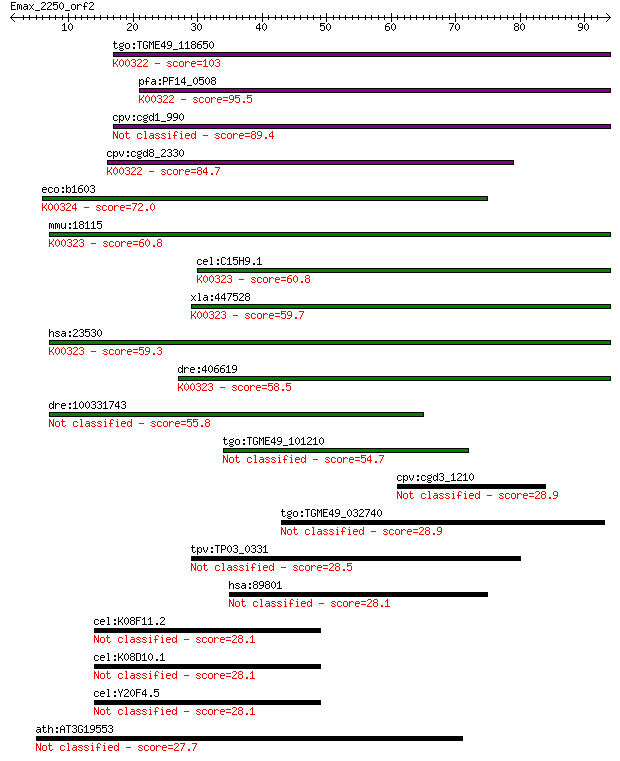
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 103 1e-22
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 95.5 4e-20
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 89.4 2e-18
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 84.7 6e-17
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 72.0 4e-13
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 60.8 1e-09
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 60.8 1e-09
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 59.7 2e-09
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 59.3 3e-09
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 58.5 5e-09
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 55.8 3e-08
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 54.7 7e-08
cpv:cgd3_1210 hypothetical protein 28.9 4.1
tgo:TGME49_032740 hypothetical protein 28.9 4.1
tpv:TP03_0331 hypothetical protein 28.5 5.1
hsa:89801 PPP1R3F, HB2E; protein phosphatase 1, regulatory (in... 28.1 7.2
cel:K08F11.2 hypothetical protein 28.1 8.1
cel:K08D10.1 hypothetical protein 28.1 8.1
cel:Y20F4.5 hypothetical protein 28.1 8.1
ath:AT3G19553 amino acid permease family protein 27.7 8.9
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 51/77 (66%), Positives = 60/77 (77%), Gaps = 0/77 (0%)
Query 17 LGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTA 76
LG+ L +EL+ TLLALSLIVGYY VW+VTPALHTPLMSVTNALSGVI+IG MLEYG +
Sbjct 923 LGVTLKALELQQFTLLALSLIVGYYSVWSVTPALHTPLMSVTNALSGVIIIGSMLEYGPS 982
Query 77 LISGFTLLAIIGTFLAS 93
S + A+ TF +S
Sbjct 983 ATSASAICALCATFFSS 999
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 95.5 bits (236), Expect = 4e-20, Method: Composition-based stats.
Identities = 44/73 (60%), Positives = 56/73 (76%), Gaps = 0/73 (0%)
Query 21 LDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISG 80
L +L+ L L LS IVGYYCVW+VTPALHTPLMS+TNALSGVI+IG M+EYG
Sbjct 1085 LSQSDLQSLFLFTLSTIVGYYCVWSVTPALHTPLMSMTNALSGVIIIGSMIEYGNGYKDL 1144
Query 81 FTLLAIIGTFLAS 93
++L+++ TFL+S
Sbjct 1145 SSILSMLATFLSS 1157
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 89.4 bits (220), Expect = 2e-18, Method: Composition-based stats.
Identities = 39/77 (50%), Positives = 58/77 (75%), Gaps = 0/77 (0%)
Query 17 LGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTA 76
LG+ + ++++++ +S ++GYYCVW V P LHTPLMSVTNALSGVI+IG M++YG
Sbjct 1052 LGLTMTTIQIQNIFSFIISTMLGYYCVWDVDPKLHTPLMSVTNALSGVIIIGSMMQYGNQ 1111
Query 77 LISGFTLLAIIGTFLAS 93
++ TL+++ TFLAS
Sbjct 1112 TVTYTTLMSMFSTFLAS 1128
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 84.7 bits (208), Expect = 6e-17, Method: Composition-based stats.
Identities = 38/63 (60%), Positives = 51/63 (80%), Gaps = 0/63 (0%)
Query 16 GLGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGT 75
GLG ++D L ++ + +LS+IVGYYC+W VTP+LHTPLMSVTNALSG+I+IG MLE G
Sbjct 1023 GLGYIIDHDTLGNILVFSLSVIVGYYCIWNVTPSLHTPLMSVTNALSGIIIIGAMLECGP 1082
Query 76 ALI 78
++
Sbjct 1083 VIL 1085
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 47/69 (68%), Gaps = 9/69 (13%)
Query 6 GWVEGVAKRCGLGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVI 65
GW+ VA + LG H T+ AL+ +VGYY VW V+ ALHTPLMSVTNA+SG+I
Sbjct 414 GWMASVAPKEFLG---------HFTVFALACVVGYYVVWNVSHALHTPLMSVTNAISGII 464
Query 66 VIGCMLEYG 74
V+G +L+ G
Sbjct 465 VVGALLQIG 473
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/95 (41%), Positives = 54/95 (56%), Gaps = 13/95 (13%)
Query 7 WVEGVAKRCGLGIVLDPMELKHL-TLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVI 65
+ G+ GLGIV + + T L+ I+GY+ VW VTPALH+PLMSVTNA+SG+
Sbjct 479 YSAGLTGMLGLGIVAPNVAFSQMVTTFGLAGIIGYHTVWGVTPALHSPLMSVTNAISGLT 538
Query 66 VIGCMLEYGTALISGF-------TLLAIIGTFLAS 93
+G G AL+ G LA + TF++S
Sbjct 539 AVG-----GLALMGGHFYPSTTSQSLAALATFISS 568
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 44/66 (66%), Gaps = 2/66 (3%)
Query 30 TLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIG--CMLEYGTALISGFTLLAII 87
T AL+ +VGY+ VW VTPALH+PLMSVTNA+SG G C++ G + +A++
Sbjct 457 TTFALAGLVGYHTVWGVTPALHSPLMSVTNAISGTTAAGALCLMGGGLMPQNSAQTMALL 516
Query 88 GTFLAS 93
TF++S
Sbjct 517 ATFISS 522
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/67 (47%), Positives = 45/67 (67%), Gaps = 2/67 (2%)
Query 29 LTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFT--LLAI 86
+T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+ +G + G + T LLA+
Sbjct 502 VTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLALMGGGYLPTNTHELLAV 561
Query 87 IGTFLAS 93
+ F++S
Sbjct 562 LAAFVSS 568
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 51/90 (56%), Gaps = 3/90 (3%)
Query 7 WVEGVAKRCGLGIVLDPMELKHL-TLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVI 65
+ G+ GLGI + + T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+
Sbjct 479 YTAGLTGILGLGIAAPNLAFSQMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLT 538
Query 66 VIGCMLEYGTALISGFTL--LAIIGTFLAS 93
+G + G L T LA + F++S
Sbjct 539 AVGGLALMGGHLYPSTTSQGLAALAAFISS 568
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 45/69 (65%), Gaps = 2/69 (2%)
Query 27 KHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFT--LL 84
+ +T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+ +G + G + T L
Sbjct 496 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLSLMGGGYLPSSTAETL 555
Query 85 AIIGTFLAS 93
A++ F++S
Sbjct 556 AVLAAFISS 564
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/59 (49%), Positives = 38/59 (64%), Gaps = 1/59 (1%)
Query 7 WVEGVAKRCGLGIVLDPMELKHL-TLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGV 64
+ G++ GLGI + T L+ IVGY+ VW VTPALH+PLMSVTNA+SG+
Sbjct 179 YTAGLSTVLGLGIASPNAAFTQMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGL 237
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/38 (65%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 34 LSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCML 71
L+ VGY VW V PALHTPLMSVTNA+SG +++G ML
Sbjct 1057 LACFVGYLLVWNVAPALHTPLMSVTNAISGTVLVGGML 1094
> cpv:cgd3_1210 hypothetical protein
Length=464
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 61 LSGVIVIGCMLEYGTALISGFTL 83
LS +I+I +L +G+AL SG TL
Sbjct 9 LSALIIITLLLSFGSALFSGLTL 31
> tgo:TGME49_032740 hypothetical protein
Length=380
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 26/50 (52%), Gaps = 0/50 (0%)
Query 43 VWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALISGFTLLAIIGTFLA 92
+W + P P+++ N L V+ M E+ TA +S + +IG+F A
Sbjct 200 LWILRPLPCLPMLTGANCLGSVLYPITMSEFTTADVSDSVMNGVIGSFPA 249
> tpv:TP03_0331 hypothetical protein
Length=806
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query 29 LTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYGTALIS 79
++++ +++++GY ++ V A +T +T+ L +I++G +++G LIS
Sbjct 462 ISMVTIAILMGYIVLFFVPYAFYTITSPMTHILE-IILLGLFVDFGNTLIS 511
> hsa:89801 PPP1R3F, HB2E; protein phosphatase 1, regulatory (inhibitor)
subunit 3F
Length=453
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 35 SLIVGYYCVWAVTPALHTPLMSVTNALSGVIVIGCMLEYG 74
+L V CV A+ P L PL L+G++V+ L G
Sbjct 398 TLAVPAECVCALPPQLRGPLTQTLGVLAGLVVVPVALNSG 437
> cel:K08F11.2 hypothetical protein
Length=886
Score = 28.1 bits (61), Expect = 8.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 14 RCGLGIVLDPMELKHLTLLALSLIVGYYCVWAVTP 48
RCG G +L ++L+H L LIVG + + P
Sbjct 583 RCGSGPILQNVKLEHYLPAMLHLIVGLFTKYIFEP 617
> cel:K08D10.1 hypothetical protein
Length=895
Score = 28.1 bits (61), Expect = 8.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 14 RCGLGIVLDPMELKHLTLLALSLIVGYYCVWAVTP 48
RCG G +L ++L+H L LIVG + + P
Sbjct 583 RCGSGPILQNVKLEHYLPAMLHLIVGLFTKYIFEP 617
> cel:Y20F4.5 hypothetical protein
Length=725
Score = 28.1 bits (61), Expect = 8.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 14 RCGLGIVLDPMELKHLTLLALSLIVGYYCVWAVTP 48
RCG G +L ++L+H L LIVG + + P
Sbjct 435 RCGSGPILQNVKLEHYLPAMLHLIVGLFTKYIFEP 469
> ath:AT3G19553 amino acid permease family protein
Length=479
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 29/66 (43%), Gaps = 0/66 (0%)
Query 5 QGWVEGVAKRCGLGIVLDPMELKHLTLLALSLIVGYYCVWAVTPALHTPLMSVTNALSGV 64
+GW++ A LG+ M LL +S I +A TP +S+ + +GV
Sbjct 285 KGWIQAAAAMSNLGLFEAEMSSDAFQLLGMSEIGMLPAFFAQRSKYGTPTISILCSATGV 344
Query 65 IVIGCM 70
I + M
Sbjct 345 IFLSWM 350
Lambda K H
0.328 0.143 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2074765676
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40