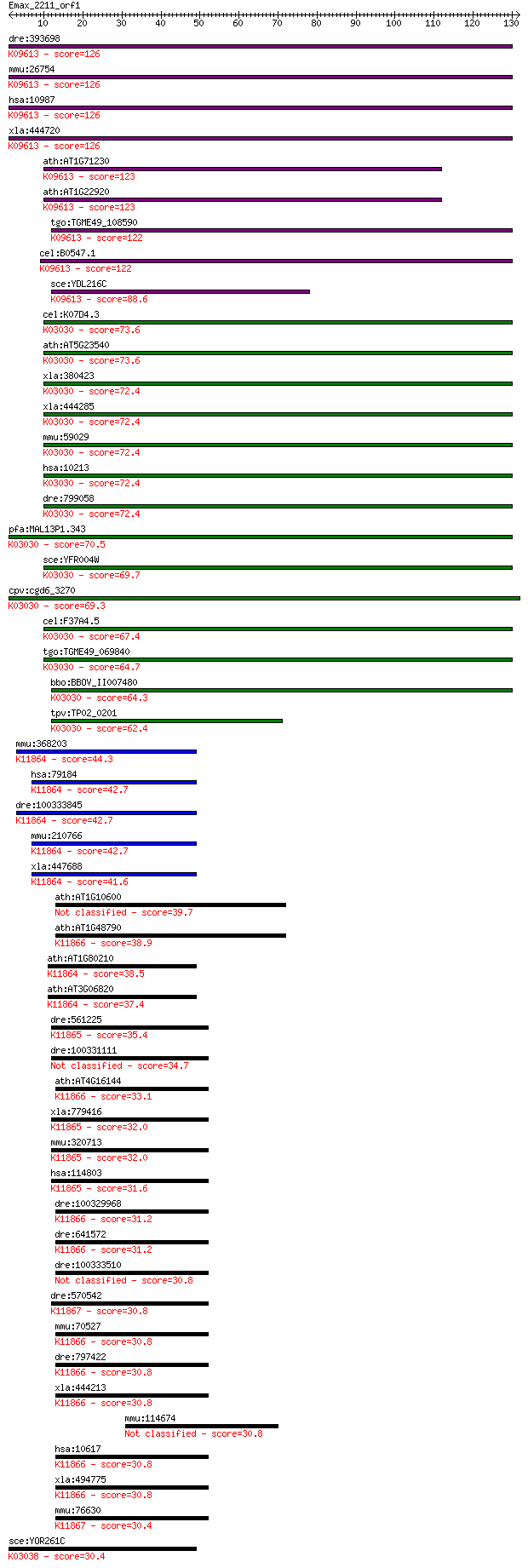
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2211_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
dre:393698 cops5, MGC73130, zgc:73130; COP9 constitutive photo... 126 2e-29
mmu:26754 Cops5, AI303502, CSN5, Jab1, Mov34, Sgn5; COP9 (cons... 126 2e-29
hsa:10987 COPS5, CSN5, JAB1, MGC3149, MOV-34, SGN5; COP9 const... 126 2e-29
xla:444720 cops5, MGC84682; COP9 constitutive photomorphogenic... 126 2e-29
ath:AT1G71230 CSN5B; CSN5B (COP9-SIGNALOSOME 5B); protein bind... 123 2e-28
ath:AT1G22920 CSN5A; CSN5A (COP9 SIGNALOSOME 5A); K09613 COP9 ... 123 2e-28
tgo:TGME49_108590 Mov34/MPN/PAD-1 domain-containing protein ; ... 122 2e-28
cel:B0547.1 csn-5; COP-9 SigNalosome subunit family member (cs... 122 4e-28
sce:YDL216C RRI1, CSN5, JAB1; Catalytic subunit of the COP9 si... 88.6 4e-18
cel:K07D4.3 rpn-11; proteasome Regulatory Particle, Non-ATPase... 73.6 2e-13
ath:AT5G23540 26S proteasome regulatory subunit, putative; K03... 73.6 2e-13
xla:380423 psmd14, MGC53911; 26S proteasome-associated pad1 ho... 72.4 3e-13
xla:444285 psmd14, MGC80929; proteasome (prosome, macropain) 2... 72.4 3e-13
mmu:59029 Psmd14, 2610312C03Rik, 3200001M20Rik, AA986732, Pad1... 72.4 3e-13
hsa:10213 PSMD14, PAD1, POH1, RPN11; proteasome (prosome, macr... 72.4 3e-13
dre:799058 psmd14, MGC162272, zgc:162272; proteasome (prosome,... 72.4 4e-13
pfa:MAL13P1.343 proteasome regulatory subunit, putative; K0303... 70.5 1e-12
sce:YFR004W RPN11, MPR1; Metalloprotease subunit of the 19S re... 69.7 2e-12
cpv:cgd6_3270 26S proteasome-associated Mov34/MPN/PAD-1 family... 69.3 3e-12
cel:F37A4.5 hypothetical protein; K03030 26S proteasome regula... 67.4 1e-11
tgo:TGME49_069840 26S proteasome non-ATPase subunit, putative ... 64.7 7e-11
bbo:BBOV_II007480 18.m06620; 26S proteasome regulatory subunit... 64.3 9e-11
tpv:TP02_0201 proteasome regulatory subunit; K03030 26S protea... 62.4 4e-10
mmu:368203 Gm5136, C6.1AL, EG368203, MGC155743, MGC155745; pre... 44.3 1e-04
hsa:79184 BRCC3, BRCC36, C6.1A, CXorf53; BRCA1/BRCA2-containin... 42.7 3e-04
dre:100333845 BRCA1/BRCA2-containing complex subunit 3-like; K... 42.7 3e-04
mmu:210766 Brcc3, C6.1A, MGC29148; BRCA1/BRCA2-containing comp... 42.7 3e-04
xla:447688 brcc3, MGC99130, brcc36; BRCA1/BRCA2-containing com... 41.6 6e-04
ath:AT1G10600 hypothetical protein 39.7 0.002
ath:AT1G48790 mov34 family protein; K11866 STAM-binding protei... 38.9 0.004
ath:AT1G80210 hypothetical protein; K11864 BRCA1/BRCA2-contain... 38.5 0.006
ath:AT3G06820 mov34 family protein; K11864 BRCA1/BRCA2-contain... 37.4 0.013
dre:561225 mysm1, im:7153144, si:ch211-59d15.8; Myb-like, SWIR... 35.4 0.046
dre:100331111 histone H2A deubiquitinase MYSM1-like 34.7 0.071
ath:AT4G16144 hypothetical protein; K11866 STAM-binding protei... 33.1 0.26
xla:779416 mysm1, 2A-DUB, MGC154322; myb-like, SWIRM and MPN d... 32.0 0.48
mmu:320713 Mysm1, C130067A03Rik, C530050H10Rik; myb-like, SWIR... 32.0 0.56
hsa:114803 MYSM1, 2A-DUB, 2ADUB, DKFZp779J1554, DKFZp779J1721,... 31.6 0.66
dre:100329968 STAM binding protein-like; K11866 STAM-binding p... 31.2 0.95
dre:641572 stambpb, MGC123247, zgc:123247; STAM binding protei... 31.2 0.98
dre:100333510 STAM binding protein-like 30.8 1.0
dre:570542 Stam binding protein like 1-like; K11867 AMSH-like ... 30.8 1.0
mmu:70527 Stambp, 5330424L14Rik, 5730422L11Rik, AW107289, Amsh... 30.8 1.1
dre:797422 stambpa, MGC66147, stambp, zgc:66147; STAM binding ... 30.8 1.1
xla:444213 stambp-b, MGC80758, amsh, stambpb; STAM binding pro... 30.8 1.1
mmu:114674 Gtf2ird2, 1700012P16Rik; GTF2I repeat domain contai... 30.8 1.2
hsa:10617 STAMBP, AMSH, MGC126516, MGC126518; STAM binding pro... 30.8 1.2
xla:494775 stambp-a, amsh, stambp, stambpa; STAM binding prote... 30.8 1.2
mmu:76630 Stambpl1, 1700095N21Rik, 8230401J17Rik, ALMalpha, AM... 30.4 1.4
sce:YOR261C RPN8; Essential, non-ATPase regulatory subunit of ... 30.4 1.5
> dre:393698 cops5, MGC73130, zgc:73130; COP9 constitutive photomorphogenic
homolog subunit 5; K09613 COP9 signalosome complex
subunit 5 [EC:3.4.-.-]
Length=334
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 80/129 (62%), Gaps = 4/129 (3%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E A++V + +GWYHSHP YGCWLSGIDV TQ L QQ Q+PF+A+V+DP RT+ +
Sbjct 120 ENAKQVGRLENAIGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKV 179
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
LG FRT P K D G +E Q +P K+ DFG H K+YY L V + +S+ L
Sbjct 180 NLGAFRT-YPKGYKPPDEGPSEY---QTIPLNKIEDFGVHCKQYYALEVSYFKSSLDRKL 235
Query 121 LEQMCSASW 129
LE + + W
Sbjct 236 LELLWNKYW 244
> mmu:26754 Cops5, AI303502, CSN5, Jab1, Mov34, Sgn5; COP9 (constitutive
photomorphogenic) homolog, subunit 5 (Arabidopsis
thaliana); K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=334
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 80/129 (62%), Gaps = 4/129 (3%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E A++V + +GWYHSHP YGCWLSGIDV TQ L QQ Q+PF+A+V+DP RT+ +
Sbjct 122 ENAKQVGRLENAIGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKV 181
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
LG FRT P K D G +E Q +P K+ DFG H K+YY L V + +S+ L
Sbjct 182 NLGAFRT-YPKGYKPPDEGPSEY---QTIPLNKIEDFGVHCKQYYALEVSYFKSSLDRKL 237
Query 121 LEQMCSASW 129
LE + + W
Sbjct 238 LELLWNKYW 246
> hsa:10987 COPS5, CSN5, JAB1, MGC3149, MOV-34, SGN5; COP9 constitutive
photomorphogenic homolog subunit 5 (Arabidopsis);
K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=334
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 80/129 (62%), Gaps = 4/129 (3%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E A++V + +GWYHSHP YGCWLSGIDV TQ L QQ Q+PF+A+V+DP RT+ +
Sbjct 122 ENAKQVGRLENAIGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKV 181
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
LG FRT P K D G +E Q +P K+ DFG H K+YY L V + +S+ L
Sbjct 182 NLGAFRT-YPKGYKPPDEGPSEY---QTIPLNKIEDFGVHCKQYYALEVSYFKSSLDRKL 237
Query 121 LEQMCSASW 129
LE + + W
Sbjct 238 LELLWNKYW 246
> xla:444720 cops5, MGC84682; COP9 constitutive photomorphogenic
homolog subunit 5; K09613 COP9 signalosome complex subunit
5 [EC:3.4.-.-]
Length=332
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/129 (47%), Positives = 80/129 (62%), Gaps = 4/129 (3%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E A++V + +GWYHSHP YGCWLSGIDV TQ L QQ Q+PF+A+V+DP RT+ +
Sbjct 120 ENAKQVGRLENAIGWYHSHPGYGCWLSGIDVSTQMLNQQFQEPFVAVVIDPTRTISAGKV 179
Query 61 ELGGFRTLLPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPL 120
LG FRT P K D G +E Q +P K+ DFG H K+YY L V + +S+ L
Sbjct 180 NLGAFRT-YPKGYKPPDEGPSEY---QTIPLNKIEDFGVHCKQYYALEVTYFKSSLDRKL 235
Query 121 LEQMCSASW 129
LE + + W
Sbjct 236 LELLWNKYW 244
> ath:AT1G71230 CSN5B; CSN5B (COP9-SIGNALOSOME 5B); protein binding;
K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=358
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 55/102 (53%), Positives = 69/102 (67%), Gaps = 4/102 (3%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP YGCWLSGIDV TQ+L QQHQ+PFLA+V+DP RTV +E+G FRT
Sbjct 135 ENVVGWYHSHPGYGCWLSGIDVSTQRLNQQHQEPFLAVVIDPTRTVSAGKVEIGAFRTYS 194
Query 70 PGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHW 111
G + D+ + Q +P K+ DFG H K+YY L V +
Sbjct 195 KGYKPPDEP----VSEYQTIPLNKIEDFGVHCKQYYSLDVTY 232
> ath:AT1G22920 CSN5A; CSN5A (COP9 SIGNALOSOME 5A); K09613 COP9
signalosome complex subunit 5 [EC:3.4.-.-]
Length=357
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 54/102 (52%), Positives = 68/102 (66%), Gaps = 4/102 (3%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP YGCWLSGIDV TQ L QQ+Q+PFLA+V+DP RTV +E+G FRT
Sbjct 135 ENVVGWYHSHPGYGCWLSGIDVSTQMLNQQYQEPFLAVVIDPTRTVSAGKVEIGAFRTYP 194
Query 70 PGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHW 111
G + DD + Q +P K+ DFG H K+YY L + +
Sbjct 195 EGHKISDD----HVSEYQTIPLNKIEDFGVHCKQYYSLDITY 232
> tgo:TGME49_108590 Mov34/MPN/PAD-1 domain-containing protein
; K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=321
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 57/119 (47%), Positives = 77/119 (64%), Gaps = 1/119 (0%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLP- 70
VVGWYHSHP Y CWLSGIDV TQ+L Q+ QDPF+A+VVDP RT+ +++G FR
Sbjct 159 VVGWYHSHPGYRCWLSGIDVETQKLHQRGQDPFVAVVVDPTRTLATGEVDIGAFRCYPDH 218
Query 71 GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLEQMCSASW 129
+ R D+ +PAEK +DFG HW+ YY+L V +S+ A L+E++ A+W
Sbjct 219 SQNARLDSVQKSRHDQAGVPAEKASDFGVHWREYYKLNVELLCSSLDALLIERLSEAAW 277
> cel:B0547.1 csn-5; COP-9 SigNalosome subunit family member (csn-5);
K09613 COP9 signalosome complex subunit 5 [EC:3.4.-.-]
Length=368
Score = 122 bits (305), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 56/121 (46%), Positives = 78/121 (64%), Gaps = 4/121 (3%)
Query 9 QQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTL 68
++KVVGWYHSHP YGCWLSGIDV TQ L Q+ Q+P++AIV+DP+RT+ +++G FRT
Sbjct 131 KEKVVGWYHSHPGYGCWLSGIDVSTQTLNQKFQEPWVAIVIDPLRTMSAGKVDIGAFRTY 190
Query 69 LPGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLEQMCSAS 128
G DD + Q +P K+ DFG H KRYY L V + + + A +L + ++
Sbjct 191 PEGYRPPDDV----PSEYQSIPLAKIEDFGVHCKRYYSLDVSFFKSQLDAHILTSLWNSY 246
Query 129 W 129
W
Sbjct 247 W 247
> sce:YDL216C RRI1, CSN5, JAB1; Catalytic subunit of the COP9
signalosome (CSN) complex that acts as an isopeptidase in cleaving
the ubiquitin-like protein Nedd8 from SCF ubiquitin ligases;
metalloendopeptidase involved in the adaptation to pheromone
signaling (EC:3.4.-.-); K09613 COP9 signalosome complex
subunit 5 [EC:3.4.-.-]
Length=440
Score = 88.6 bits (218), Expect = 4e-18, Method: Composition-based stats.
Identities = 36/66 (54%), Positives = 50/66 (75%), Gaps = 3/66 (4%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLPG 71
VVGW+HSHP Y CWLS ID+ TQ L Q+ QDP++AIVVDP++++ ++ + +G FRT+
Sbjct 159 VVGWFHSHPGYDCWLSNIDIQTQDLNQRFQDPYVAIVVDPLKSLEDKILRMGAFRTI--- 215
Query 72 EEKRDD 77
E K DD
Sbjct 216 ESKSDD 221
> cel:K07D4.3 rpn-11; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-11); K03030 26S proteasome regulatory
subunit N11
Length=312
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 68/128 (53%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP +GCWLSG+D+ TQQ + D +A+VVDPI++V + + + FRT+
Sbjct 108 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSDRAVAVVVDPIQSVKGKVV-IDAFRTIN 166
Query 70 P-----GEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
P +E R + G + S Q L ++ H YY + + + + + +L
Sbjct 167 PQSMALNQEPRQTTSNLGHLQKPSIQAL----IHGLNRH---YYSIPIAYRTHDLEQKML 219
Query 122 EQMCSASW 129
+ SW
Sbjct 220 LNLNKLSW 227
> ath:AT5G23540 26S proteasome regulatory subunit, putative; K03030
26S proteasome regulatory subunit N11
Length=259
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + +A+VVDPI++V + + + FR
Sbjct 56 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALNQRAVAVVVDPIQSVKGKVV-IDAFRSIN 114
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
T++ G+E R + G S Q L ++ H YY + +++ N + +L
Sbjct 115 PQTIMLGQEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSIAINYRKNELEEKML 167
Query 122 EQMCSASW 129
+ W
Sbjct 168 LNLHKKKW 175
> xla:380423 psmd14, MGC53911; 26S proteasome-associated pad1
homolog (EC:3.1.2.15); K03030 26S proteasome regulatory subunit
N11
Length=310
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR
Sbjct 106 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 164
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
++ G E R + G S Q L ++ H YY + +++ N + +L
Sbjct 165 ANMMVLGHEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSITINYRKNELEQKML 217
Query 122 EQMCSASW 129
+ SW
Sbjct 218 LNLHKKSW 225
> xla:444285 psmd14, MGC80929; proteasome (prosome, macropain)
26S subunit, non-ATPase, 14; K03030 26S proteasome regulatory
subunit N11
Length=310
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR
Sbjct 106 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 164
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
++ G E R + G S Q L ++ H YY + +++ N + +L
Sbjct 165 ANMMVLGHEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSITINYRKNELEQKML 217
Query 122 EQMCSASW 129
+ SW
Sbjct 218 LNLHKKSW 225
> mmu:59029 Psmd14, 2610312C03Rik, 3200001M20Rik, AA986732, Pad1,
Poh1, rpm11; proteasome (prosome, macropain) 26S subunit,
non-ATPase, 14 (EC:3.1.2.15); K03030 26S proteasome regulatory
subunit N11
Length=310
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR
Sbjct 106 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 164
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
++ G E R + G S Q L ++ H YY + +++ N + +L
Sbjct 165 ANMMVLGHEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSITINYRKNELEQKML 217
Query 122 EQMCSASW 129
+ SW
Sbjct 218 LNLHKKSW 225
> hsa:10213 PSMD14, PAD1, POH1, RPN11; proteasome (prosome, macropain)
26S subunit, non-ATPase, 14 (EC:3.1.2.15); K03030 26S
proteasome regulatory subunit N11
Length=310
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR
Sbjct 106 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 164
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
++ G E R + G S Q L ++ H YY + +++ N + +L
Sbjct 165 ANMMVLGHEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSITINYRKNELEQKML 217
Query 122 EQMCSASW 129
+ SW
Sbjct 218 LNLHKKSW 225
> dre:799058 psmd14, MGC162272, zgc:162272; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 14 (EC:3.1.2.15); K03030
26S proteasome regulatory subunit N11
Length=310
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 16/128 (12%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFR--- 66
+ VVGWYHSHP +GCWLSG+D+ TQQ + + +A+VVDPI++V + + + FR
Sbjct 106 EMVVGWYHSHPGFGCWLSGVDINTQQSFEALSERAVAVVVDPIQSVKGKVV-IDAFRLIN 164
Query 67 --TLLPGEEKR---DDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLL 121
++ G E R + G S Q L ++ H YY + +++ N + +L
Sbjct 165 ANMMVLGHEPRQTTSNLGHLNKPSIQAL----IHGLNRH---YYSITINYRKNELEQKML 217
Query 122 EQMCSASW 129
+ SW
Sbjct 218 LNLHKKSW 225
> pfa:MAL13P1.343 proteasome regulatory subunit, putative; K03030
26S proteasome regulatory subunit N11
Length=311
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 42/134 (31%), Positives = 67/134 (50%), Gaps = 10/134 (7%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E +K + VVGWYHSHP +GCWLSG DV TQ+ +Q + +VVDPI++V + +
Sbjct 99 EELKKTGRHEMVVGWYHSHPGFGCWLSGTDVNTQKSFEQLNPRTIGVVVDPIQSVKGKVV 158
Query 61 ELGGFRTLLP-----GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANS 115
+ FR + P G+E R + + L A V+ + YY + +++ N
Sbjct 159 -IDCFRLINPHILMLGQEPRQTTSNIGYLTKPTLTA-LVHGLN---RNYYSIVINYRKNE 213
Query 116 ISAPLLEQMCSASW 129
+ +L + W
Sbjct 214 LEKNMLLNLHKDMW 227
> sce:YFR004W RPN11, MPR1; Metalloprotease subunit of the 19S
regulatory particle of the 26S proteasome lid; couples the deubiquitination
and degradation of proteasome substrates; involved,
independent of catalytic activity, in fission of mitochondria
and peroxisomes; K03030 26S proteasome regulatory
subunit N11
Length=306
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 38/121 (31%), Positives = 60/121 (49%), Gaps = 2/121 (1%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
Q VVGWYHSHP +GCWLS +DV TQ+ +Q +A+VVDPI++V + + + FR +
Sbjct 102 QMVVGWYHSHPGFGCWLSSVDVNTQKSFEQLNSRAVAVVVDPIQSVKGKVV-IDAFRLID 160
Query 70 PGEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKR-YYQLRVHWCANSISAPLLEQMCSAS 128
G + +++ LL + R YY L + + + +L +
Sbjct 161 TGALINNLEPRQTTSNTGLLNKANIQALIHGLNRHYYSLNIDYHKTAKETKMLMNLHKEQ 220
Query 129 W 129
W
Sbjct 221 W 221
> cpv:cgd6_3270 26S proteasome-associated Mov34/MPN/PAD-1 family.
JAB domain. ; K03030 26S proteasome regulatory subunit N11
Length=315
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 67/136 (49%), Gaps = 10/136 (7%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSI 60
E ++V + VVGWYHSHP +GCW SG DV TQQ +Q + IVVDPI++V + +
Sbjct 103 EMLKRVGRSELVVGWYHSHPGFGCWFSGTDVSTQQSFEQLNPRAVGIVVDPIQSVKGKVV 162
Query 61 ELGGFRTLLP-----GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANS 115
+ FR + P G+E R + P+ G + + YY + + + N
Sbjct 163 -IDCFRLISPHSVIAGQEPRQTTSNIGHLQK---PSITALVHGLN-RNYYSIAIRYRKNL 217
Query 116 ISAPLLEQMCSASWPQ 131
+ +L + +W +
Sbjct 218 LEQKMLLNLHKPTWSE 233
> cel:F37A4.5 hypothetical protein; K03030 26S proteasome regulatory
subunit N11
Length=319
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/133 (33%), Positives = 65/133 (48%), Gaps = 21/133 (15%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP +GCWLS +DV TQQ + +A+VVDPI++V + + L FR++
Sbjct 104 ENVVGWYHSHPGFGCWLSSVDVNTQQSFEALHPRAVAVVVDPIQSVKGK-VMLDAFRSVN 162
Query 70 P-GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWK------------RYYQLRVHWCANSI 116
P + R A AE P + ++ G K +YY L V + S
Sbjct 163 PLNLQIRPLAPTAE-------PRQTTSNLGHLTKPSLISVVHGLGTKYYSLNVAYRMGSN 215
Query 117 SAPLLEQMCSASW 129
+L + SW
Sbjct 216 EQKMLMCLNKKSW 228
> tgo:TGME49_069840 26S proteasome non-ATPase subunit, putative
; K03030 26S proteasome regulatory subunit N11
Length=314
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 62/125 (49%), Gaps = 10/125 (8%)
Query 10 QKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+ VVGWYHSHP +GCW SG DV TQQ +Q + +VVDPI++V + + + FR +
Sbjct 111 EMVVGWYHSHPGFGCWFSGTDVNTQQSFEQLNPRAVGVVVDPIQSVKGKVV-IDCFRLIN 169
Query 70 P-----GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLEQM 124
P G+E R + + A V+ + YY + +++ N + +L +
Sbjct 170 PHLLMLGQELRQTTSNIGHLQRPTISA-LVHGLN---RNYYAIVINYRKNELENQMLLNL 225
Query 125 CSASW 129
W
Sbjct 226 HRNKW 230
> bbo:BBOV_II007480 18.m06620; 26S proteasome regulatory subunit;
K03030 26S proteasome regulatory subunit N11
Length=312
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 59/123 (47%), Gaps = 10/123 (8%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLP- 70
VVGWYHSHP +GCW SG D+ TQQ +Q + IV+DPI++V + + + FR + P
Sbjct 111 VVGWYHSHPGFGCWFSGTDINTQQSFEQLNPRAVGIVIDPIQSVKGKVV-IDCFRLITPH 169
Query 71 ----GEEKRDDAGDAEAASSQLLPAEKVNDFGAHWKRYYQLRVHWCANSISAPLLEQMCS 126
G+E R + S + A + YY + +++ + + +L
Sbjct 170 LIMLGQEPRQTTSNIGHLSKPTMIAV----VHGLNRNYYNIVINYRKSVLETQMLMNYHR 225
Query 127 ASW 129
W
Sbjct 226 NKW 228
> tpv:TP02_0201 proteasome regulatory subunit; K03030 26S proteasome
regulatory subunit N11
Length=312
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 39/59 (66%), Gaps = 1/59 (1%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLP 70
VVGWYHSHP +GCW SG DV TQQ +Q + +V+DPI++V + + + FR + P
Sbjct 111 VVGWYHSHPGFGCWFSGTDVNTQQSFEQLNPRAVGVVIDPIQSVKGKVV-IDCFRLISP 168
> mmu:368203 Gm5136, C6.1AL, EG368203, MGC155743, MGC155745; predicted
gene 5136 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=291
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 3 AEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
AE+ +VVGWYHSHP+ W S +DV TQ + Q F+ ++
Sbjct 108 AEQTGRPMRVVGWYHSHPHITVWPSHVDVRTQAMYQMMDQSFVGLI 153
> hsa:79184 BRCC3, BRCC36, C6.1A, CXorf53; BRCA1/BRCA2-containing
complex, subunit 3 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=291
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 7 PMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
PM +VVGWYHSHP+ W S +DV TQ + Q F+ ++
Sbjct 114 PM--RVVGWYHSHPHITVWPSHVDVRTQAMYQMMDQGFVGLI 153
> dre:100333845 BRCA1/BRCA2-containing complex subunit 3-like;
K11864 BRCA1/BRCA2-containing complex subunit 3
Length=260
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 3 AEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
AE +VVGWYHSHP+ W S +DV TQ + Q F+ ++
Sbjct 78 AEMTGRPMRVVGWYHSHPHITVWPSHVDVRTQAMYQMMDQGFVGLI 123
> mmu:210766 Brcc3, C6.1A, MGC29148; BRCA1/BRCA2-containing complex,
subunit 3 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=291
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 7 PMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
PM +VVGWYHSHP+ W S +DV TQ + Q F+ ++
Sbjct 114 PM--RVVGWYHSHPHITVWPSHVDVRTQAMYQMMDQGFVGLI 153
> xla:447688 brcc3, MGC99130, brcc36; BRCA1/BRCA2-containing complex,
subunit 3 (EC:3.1.2.15); K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=261
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 7 PMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
PM +VVGWYHSHP+ W S +DV TQ + Q F+ ++
Sbjct 84 PM--RVVGWYHSHPHITVWPSHVDVRTQAMYQMMDVGFVGLI 123
> ath:AT1G10600 hypothetical protein
Length=166
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 4/59 (6%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLPG 71
VGW H+HP+ GC++S +D+ T Q AIVV P + S G F+ PG
Sbjct 66 VGWIHTHPSQGCFMSSVDLHTHYSYQVMVPEAFAIVVAP----TDSSKSYGIFKLTDPG 120
> ath:AT1G48790 mov34 family protein; K11866 STAM-binding protein
[EC:3.1.2.15]
Length=507
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 29/59 (49%), Gaps = 4/59 (6%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLLPG 71
+GW H+HP C++S IDV T Q +AIV+ P + N I FR PG
Sbjct 407 LGWIHTHPTQSCFMSSIDVHTHYSYQIMLPEAVAIVMAPQDSSRNHGI----FRLTTPG 461
> ath:AT1G80210 hypothetical protein; K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=371
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 11 KVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
+V+GWYHSHP+ S +DV TQ + Q F+ ++
Sbjct 91 RVIGWYHSHPHITVLPSHVDVRTQAMYQLLDSGFIGLI 128
> ath:AT3G06820 mov34 family protein; K11864 BRCA1/BRCA2-containing
complex subunit 3
Length=405
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 11 KVVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIV 48
+V+GWYHSHP+ S +DV TQ + Q F+ ++
Sbjct 91 RVIGWYHSHPHITVLPSHVDVRTQAMYQLLDSGFIGLI 128
> dre:561225 mysm1, im:7153144, si:ch211-59d15.8; Myb-like, SWIRM
and MPN domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=822
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQD----PFLAIVVDP 51
VVGWYHSHP + S D+ TQ Q + PF+ ++V P
Sbjct 591 VVGWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAPFIGMIVSP 634
> dre:100331111 histone H2A deubiquitinase MYSM1-like
Length=2360
Score = 34.7 bits (78), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQD----PFLAIVVDP 51
VVGWYHSHP + S D+ TQ Q + PF+ ++V P
Sbjct 2129 VVGWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAPFIGMIVSP 2172
> ath:AT4G16144 hypothetical protein; K11866 STAM-binding protein
[EC:3.1.2.15]
Length=507
Score = 33.1 bits (74), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP C++S +D+ T Q +AIV+ P
Sbjct 407 LGWIHTHPTQTCFMSSVDLHTHYSYQIMLPEAVAIVMAP 445
> xla:779416 mysm1, 2A-DUB, MGC154322; myb-like, SWIRM and MPN
domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=818
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 4/44 (9%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQD----PFLAIVVDP 51
V+GWYHSHP + S D+ TQ Q + FL +++ P
Sbjct 622 VIGWYHSHPAFDPNPSIRDIDTQAKYQNYFSRGGAKFLGMIISP 665
> mmu:320713 Mysm1, C130067A03Rik, C530050H10Rik; myb-like, SWIRM
and MPN domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=819
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 4/44 (9%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQD----PFLAIVVDP 51
V+GWYHSHP + S D+ TQ Q + F+ ++V P
Sbjct 642 VIGWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAKFIGMIVSP 685
> hsa:114803 MYSM1, 2A-DUB, 2ADUB, DKFZp779J1554, DKFZp779J1721,
KIAA1915, RP4-592A1.1; Myb-like, SWIRM and MPN domains 1
(EC:3.1.2.15); K11865 protein MYSM1
Length=828
Score = 31.6 bits (70), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 4/44 (9%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQD----PFLAIVVDP 51
V+GWYHSHP + S D+ TQ Q + F+ ++V P
Sbjct 651 VIGWYHSHPAFDPNPSLRDIDTQAKYQSYFSRGGAKFIGMIVSP 694
> dre:100329968 STAM binding protein-like; K11866 STAM-binding
protein [EC:3.1.2.15]
Length=418
Score = 31.2 bits (69), Expect = 0.95, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 325 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 363
> dre:641572 stambpb, MGC123247, zgc:123247; STAM binding protein
b (EC:3.1.2.15); K11866 STAM-binding protein [EC:3.1.2.15]
Length=418
Score = 31.2 bits (69), Expect = 0.98, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 325 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 363
> dre:100333510 STAM binding protein-like
Length=424
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 331 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 369
> dre:570542 Stam binding protein like 1-like; K11867 AMSH-like
protease [EC:3.1.2.15]
Length=420
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 12 VVGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 326 TLGWIHTHPTQTAFLSSVDLHTHSSYQLMLPEAIAIVCAP 365
> mmu:70527 Stambp, 5330424L14Rik, 5730422L11Rik, AW107289, Amsh,
KIAA4198, mKIAA4198; STAM binding protein (EC:3.1.2.15);
K11866 STAM-binding protein [EC:3.1.2.15]
Length=424
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 331 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 369
> dre:797422 stambpa, MGC66147, stambp, zgc:66147; STAM binding
protein a (EC:3.1.2.15); K11866 STAM-binding protein [EC:3.1.2.15]
Length=418
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 325 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 363
> xla:444213 stambp-b, MGC80758, amsh, stambpb; STAM binding protein
(EC:3.1.2.15); K11866 STAM-binding protein [EC:3.1.2.15]
Length=416
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 323 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 361
> mmu:114674 Gtf2ird2, 1700012P16Rik; GTF2I repeat domain containing
2
Length=936
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 3/39 (7%)
Query 31 VCTQQLQQQHQDPFLAIVVDPIRTVCNRSIELGGFRTLL 69
+C Q+L+ H + +VVD + +C+R + G F TLL
Sbjct 654 LCAQKLRMGH---VMDVVVDSVNWICSRGLNHGDFTTLL 689
> hsa:10617 STAMBP, AMSH, MGC126516, MGC126518; STAM binding protein
(EC:3.1.2.15); K11866 STAM-binding protein [EC:3.1.2.15]
Length=424
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 331 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESVAIVCSP 369
> xla:494775 stambp-a, amsh, stambp, stambpa; STAM binding protein
(EC:3.1.2.15); K11866 STAM-binding protein [EC:3.1.2.15]
Length=416
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 323 LGWIHTHPTQTAFLSSVDLHTHCSYQMMLPESIAIVCSP 361
> mmu:76630 Stambpl1, 1700095N21Rik, 8230401J17Rik, ALMalpha,
AMSH-FP; STAM binding protein like 1 (EC:3.1.2.15); K11867 AMSH-like
protease [EC:3.1.2.15]
Length=436
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 13 VGWYHSHPNYGCWLSGIDVCTQQLQQQHQDPFLAIVVDP 51
+GW H+HP +LS +D+ T Q +AIV P
Sbjct 343 LGWIHTHPTQTAFLSSVDLHTHCSYQLMLPEAIAIVCSP 381
> sce:YOR261C RPN8; Essential, non-ATPase regulatory subunit of
the 26S proteasome; has similarity to the human p40 proteasomal
subunit and to another S. cerevisiae regulatory subunit,
Rpn11p; K03038 26S proteasome regulatory subunit N8
Length=338
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 28/50 (56%), Gaps = 6/50 (12%)
Query 1 EAAEKVPMQQKVVGWYHSHPNYGCWLSGIDVCTQQLQQQH--QDPFLAIV 48
E +K+ ++K++GWYHS P L D+ +L +++ +P L IV
Sbjct 78 EMCKKINAKEKLIGWYHSGPK----LRASDLKINELFKKYTQNNPLLLIV 123
Lambda K H
0.319 0.133 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40