bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2196_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
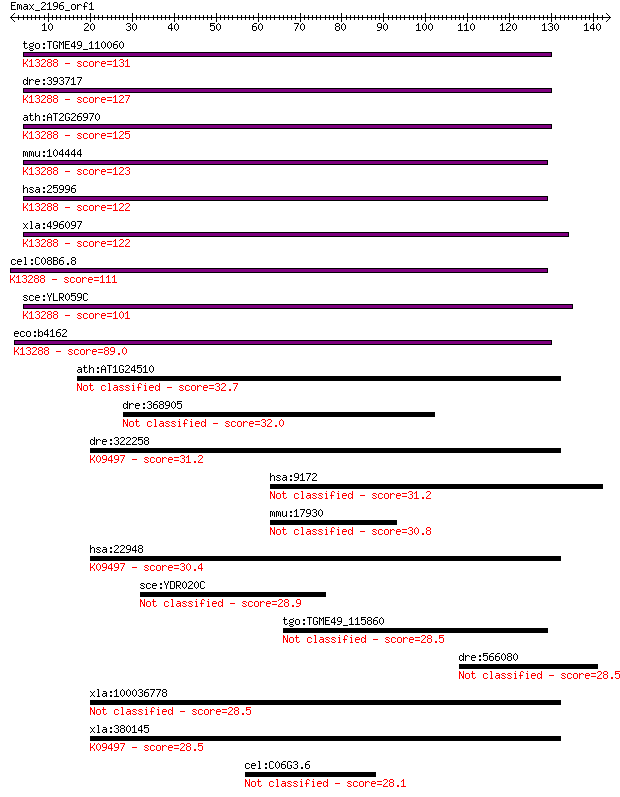
tgo:TGME49_110060 oligoribonuclease, putative ; K13288 oligori... 131 6e-31
dre:393717 smfn, MGC73191, zgc:73191; small fragment nuclease;... 127 8e-30
ath:AT2G26970 exonuclease family protein; K13288 oligoribonucl... 125 5e-29
mmu:104444 Rexo2, 1810038D15Rik, AW107347, Sfn, Smfn; REX2, RN... 123 2e-28
hsa:25996 REXO2, DKFZp566E144, MGC111570, REX2, RFN, SFN; REX2... 122 4e-28
xla:496097 rexo2, smfn; REX2, RNA exonuclease 2 homolog; K1328... 122 5e-28
cel:C08B6.8 hypothetical protein; K13288 oligoribonuclease [EC... 111 8e-25
sce:YLR059C REX2, YNT20; 3'-5' RNA exonuclease; involved in 3'... 101 7e-22
eco:b4162 orn, ECK4158, JW5740, yjeR; oligoribonuclease (EC:3.... 89.0 5e-18
ath:AT1G24510 T-complex protein 1 epsilon subunit, putative / ... 32.7 0.42
dre:368905 rab3gap2, MGC158161, RAB3, RAB3GAP, si:ch211-214p16... 32.0 0.70
dre:322258 cct5, wu:fb54h08; chaperonin containing TCP1, subun... 31.2 0.99
hsa:9172 MYOM2, TTNAP; myomesin (M-protein) 2, 165kDa 31.2 1.2
mmu:17930 Myom2, AW146149, MGC141540, MGC141541; myomesin 2 30.8
hsa:22948 CCT5, CCT-epsilon, CCTE, KIAA0098, TCP-1-epsilon; ch... 30.4 2.0
sce:YDR020C DAS2, RRT3; Das2p (EC:2.7.1.48) 28.9 5.0
tgo:TGME49_115860 EF hand domain-containing protein 28.5 6.8
dre:566080 myh11; myosin, heavy chain 11, smooth muscle 28.5 6.9
xla:100036778 cct5b; chaperonin containing TCP1, subunit 5 (ep... 28.5 7.8
xla:380145 cct5, MGC53061, cct5a; chaperonin containing TCP1, ... 28.5 7.9
cel:C06G3.6 hypothetical protein 28.1 8.9
> tgo:TGME49_110060 oligoribonuclease, putative ; K13288 oligoribonuclease
[EC:3.1.-.-]
Length=367
Score = 131 bits (330), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 61/126 (48%), Positives = 88/126 (69%), Gaps = 0/126 (0%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L+ M WC++ HG+SGLT AC +S +T +DAE++I+ F++ + + A L GNSVH+D+
Sbjct 239 LDGMDEWCKKTHGESGLTAACRKSTLTLADAEEQILEFVQKHVATTRVAPLGGNSVHVDR 298
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYY 123
+FL ++MP LI L YRI+D+S+IK +A W P + KK HRAL DI ESI+EL YY
Sbjct 299 QFLVKQMPRLIAHLSYRIIDVSTIKELAMRWKPELPLVKKAANHRALEDIHESIDELLYY 358
Query 124 RKQIFR 129
+ +FR
Sbjct 359 TRHLFR 364
> dre:393717 smfn, MGC73191, zgc:73191; small fragment nuclease;
K13288 oligoribonuclease [EC:3.1.-.-]
Length=199
Score = 127 bits (320), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 63/131 (48%), Positives = 88/131 (67%), Gaps = 9/131 (6%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLN---GVGAKEAVLAGNSVH 60
L+ M WC+EHHGKSGLTQA S +T AE ++F++ + GV LAGNSVH
Sbjct 56 LDGMSDWCKEHHGKSGLTQAVRDSHITLQQAEYEFLSFIRQHTPPGV----CPLAGNSVH 111
Query 61 MDKEFLRREMPELIEFLHYRILDISSIKVVAKSWFPR--VAPPKKRYRHRALADIQESIE 118
DK+FL + MP+ + LHYRI+D+S+IK +++ W+P P+K+ HRAL DIQESI+
Sbjct 112 ADKKFLDKYMPQFMHHLHYRIIDVSTIKELSRRWYPEEYNLAPQKKSSHRALDDIQESIK 171
Query 119 ELAYYRKQIFR 129
EL +YR +F+
Sbjct 172 ELQFYRTSVFK 182
> ath:AT2G26970 exonuclease family protein; K13288 oligoribonuclease
[EC:3.1.-.-]
Length=218
Score = 125 bits (313), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 60/128 (46%), Positives = 87/128 (67%), Gaps = 3/128 (2%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L++M WC+ HHG SGLT+ L S +T +AEQ++I F+K VG+ +LAGNSV++D
Sbjct 89 LDKMDDWCQTHHGASGLTKKVLLSAITEREAEQKVIEFVK-KHVGSGNPLLAGNSVYVDF 147
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFP--RVAPPKKRYRHRALADIQESIEELA 121
FL++ MPEL + ++D+SS+K + WFP R P K+ HRA+ DI+ESI+EL
Sbjct 148 LFLKKYMPELAALFPHILVDVSSVKALCARWFPIERRKAPAKKNNHRAMDDIRESIKELK 207
Query 122 YYRKQIFR 129
YY+K IF+
Sbjct 208 YYKKTIFK 215
> mmu:104444 Rexo2, 1810038D15Rik, AW107347, Sfn, Smfn; REX2,
RNA exonuclease 2 homolog (S. cerevisiae); K13288 oligoribonuclease
[EC:3.1.-.-]
Length=237
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 82/127 (64%), Gaps = 3/127 (2%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L+ M WC+EHHGKSGLT+A S +T AE ++F++ LAGNSVH DK
Sbjct 90 LDSMSDWCKEHHGKSGLTKAVKESTVTLQQAEYEFLSFVRQQ-TPPGLCPLAGNSVHADK 148
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVA--PPKKRYRHRALADIQESIEELA 121
+FL + MP+ ++ LHYRI+D+S++K + + W+P PKK HRAL DI ESI+EL
Sbjct 149 KFLDKHMPQFMKHLHYRIIDVSTVKELCRRWYPEDYEFAPKKAASHRALDDISESIKELQ 208
Query 122 YYRKQIF 128
+YR IF
Sbjct 209 FYRNNIF 215
> hsa:25996 REXO2, DKFZp566E144, MGC111570, REX2, RFN, SFN; REX2,
RNA exonuclease 2 homolog (S. cerevisiae); K13288 oligoribonuclease
[EC:3.1.-.-]
Length=237
Score = 122 bits (306), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 82/127 (64%), Gaps = 3/127 (2%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L+ M WC+EHHGKSGLT+A S +T AE ++F++ LAGNSVH DK
Sbjct 90 LDSMSDWCKEHHGKSGLTKAVKESTITLQQAEYEFLSFVRQQ-TPPGLCPLAGNSVHEDK 148
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVA--PPKKRYRHRALADIQESIEELA 121
+FL + MP+ ++ LHYRI+D+S++K + + W+P PKK HRAL DI ESI+EL
Sbjct 149 KFLDKYMPQFMKHLHYRIIDVSTVKELCRRWYPEEYEFAPKKAASHRALDDISESIKELQ 208
Query 122 YYRKQIF 128
+YR IF
Sbjct 209 FYRNNIF 215
> xla:496097 rexo2, smfn; REX2, RNA exonuclease 2 homolog; K13288
oligoribonuclease [EC:3.1.-.-]
Length=186
Score = 122 bits (305), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 64/133 (48%), Positives = 86/133 (64%), Gaps = 5/133 (3%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L+ M WC+EHHGKSGLTQA S +T AE ++F++ LAGNSVH DK
Sbjct 56 LDGMSDWCKEHHGKSGLTQAVRESRITLQQAEYEFLSFIR-QHTPPGVCPLAGNSVHADK 114
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFP---RVAPPKKRYRHRALADIQESIEEL 120
+FL + MP + LHYRI+D+S++K + + W P R+A PKK HRAL DI+ESI+EL
Sbjct 115 KFLDKYMPRFMRHLHYRIIDVSTVKELCRRWHPVEYRLA-PKKAASHRALDDIRESIKEL 173
Query 121 AYYRKQIFRDLED 133
+YR+ IF+ D
Sbjct 174 QFYRESIFKTKTD 186
> cel:C08B6.8 hypothetical protein; K13288 oligoribonuclease [EC:3.1.-.-]
Length=193
Score = 111 bits (277), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 54/128 (42%), Positives = 82/128 (64%), Gaps = 1/128 (0%)
Query 1 KKELEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVH 60
K+ L+ M W R ++GL + + S + +DAE +I FLKL+ + K + AGNS++
Sbjct 59 KEVLDNMEEWPRNTFHENGLMEKIIASKYSMADAENEVIDFLKLHALPGKSPI-AGNSIY 117
Query 61 MDKEFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEEL 120
MD+ F+++ MP+L +F HYR +D+S+IK + + W+P PKK+ HRA DI ESI EL
Sbjct 118 MDRLFIKKYMPKLDKFAHYRCIDVSTIKGLVQRWYPDYKHPKKQCTHRAFDDIMESIAEL 177
Query 121 AYYRKQIF 128
YR+ IF
Sbjct 178 KNYRESIF 185
> sce:YLR059C REX2, YNT20; 3'-5' RNA exonuclease; involved in
3'-end processing of U4 and U5 snRNAs, 5S and 5.8S rRNAs, and
RNase P and RNase MRP RNA; localized to mitochondria and null
suppresses escape of mtDNA to nucleus in yme1 yme2 mutants;
RNase D exonuclease (EC:3.1.-.-); K13288 oligoribonuclease
[EC:3.1.-.-]
Length=269
Score = 101 bits (252), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 53/133 (39%), Positives = 78/133 (58%), Gaps = 2/133 (1%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
+ +M WC EHHG SGLT L S T + E ++ +++ VLAGNSVHMD+
Sbjct 109 MNKMNEWCIEHHGNSGLTAKVLASEKTLAQVEDELLEYIQRYIPDKNVGVLAGNSVHMDR 168
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAP--PKKRYRHRALADIQESIEELA 121
F+ RE P++I+ L YRI+D+SSI VA+ P + PKK H A +DI+ESI +L
Sbjct 169 LFMVREFPKVIDHLFYRIVDVSSIMEVARRHNPALQARNPKKEAAHTAYSDIKESIAQLQ 228
Query 122 YYRKQIFRDLEDS 134
+Y + +++
Sbjct 229 WYMDNYLKPPQET 241
> eco:b4162 orn, ECK4158, JW5740, yjeR; oligoribonuclease (EC:3.1.-.-);
K13288 oligoribonuclease [EC:3.1.-.-]
Length=181
Score = 89.0 bits (219), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 49/129 (37%), Positives = 74/129 (57%), Gaps = 2/129 (1%)
Query 2 KELEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHM 61
++L M W H SGL + S M +AE + FLK V A ++ + GNS+
Sbjct 53 EQLALMDDWNVRTHTASGLVERVKASTMGDREAELATLEFLK-QWVPAGKSPICGNSIGQ 111
Query 62 DKEFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPP-KKRYRHRALADIQESIEEL 120
D+ FL + MPEL + HYR LD+S++K +A+ W P + K+ H+A+ DI+ES+ EL
Sbjct 112 DRRFLFKYMPELEAYFHYRYLDVSTLKELARRWKPEILDGFTKQGTHQAMDDIRESVAEL 171
Query 121 AYYRKQIFR 129
AYYR+ +
Sbjct 172 AYYREHFIK 180
> ath:AT1G24510 T-complex protein 1 epsilon subunit, putative
/ TCP-1-epsilon, putative / chaperonin, putative
Length=535
Score = 32.7 bits (73), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 51/119 (42%), Gaps = 16/119 (13%)
Query 17 KSGLTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPE 72
K L + +++ + +D E+R + +K+ G VG K E + +DK+ +MP+
Sbjct 178 KRSLAEIAVKAVLAVADLERRDVNLDLIKVEGKVGGKLEDTELIYGILIDKDMSHPQMPK 237
Query 73 LIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IE H IL PPK + +H+ D E E L +Q F ++
Sbjct 238 QIEDAHIAILTCP------------FEPPKPKTKHKVDIDTVEKFETLRKQEQQYFDEM 284
> dre:368905 rab3gap2, MGC158161, RAB3, RAB3GAP, si:ch211-214p16.5,
si:zc214p16.5, wu:fb94b08, zgc:158161; RAB3 GTPase activating
protein subunit 2 (non-catalytic)
Length=1270
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 38/75 (50%), Gaps = 1/75 (1%)
Query 28 CMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDKEFLRREMPELIEFLHYRILDISSI 87
C++ QR+ A L+ +G+ ++ + +V ++KE + P+ + L + +SSI
Sbjct 700 CLSGQSPLQRVCATLQESGISPQQLLSLLLTVWLNKEKDVLKCPDAVSHLKTLLSTLSSI 759
Query 88 K-VVAKSWFPRVAPP 101
K V +SW + P
Sbjct 760 KGAVDESWDGQCVSP 774
> dre:322258 cct5, wu:fb54h08; chaperonin containing TCP1, subunit
5 (epsilon); K09497 T-complex protein 1 subunit epsilon
Length=541
Score = 31.2 bits (69), Expect = 0.99, Method: Composition-based stats.
Identities = 28/116 (24%), Positives = 52/116 (44%), Gaps = 16/116 (13%)
Query 20 LTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPELIE 75
+ + + + +T +D E++ + F +K+ G VG K E V +DKEF +MP++++
Sbjct 185 MAEIAVNAILTVADMERKDVDFELIKVEGKVGGKLEDTQLIKGVIVDKEFSHPQMPKVLK 244
Query 76 FLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IL PPK + +H+ E + L Y K F+++
Sbjct 245 DTKIAILTCP------------FEPPKPKTKHKLDVTSVEDYKALQKYEKDKFQEM 288
> hsa:9172 MYOM2, TTNAP; myomesin (M-protein) 2, 165kDa
Length=1465
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 38/80 (47%), Gaps = 6/80 (7%)
Query 63 KEFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALA-DIQESIEELA 121
KEFLR++ P E+LH+ + + +++V K VA KK + L D+ E L
Sbjct 1122 KEFLRKQGPHFAEYLHWDVTEECEVRLVCK-----VANTKKETVFKWLKDDVLYETETLP 1176
Query 122 YYRKQIFRDLEDSLTSRDNA 141
+ I L L+ +D+
Sbjct 1177 NLERGICELLIPKLSKKDHG 1196
> mmu:17930 Myom2, AW146149, MGC141540, MGC141541; myomesin 2
Length=1463
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 63 KEFLRREMPELIEFLHYRILDISSIKVVAK 92
KEFLR++ P E+LH+ + + +++V K
Sbjct 1122 KEFLRKQGPHFAEYLHWDVTEECEVRLVCK 1151
> hsa:22948 CCT5, CCT-epsilon, CCTE, KIAA0098, TCP-1-epsilon;
chaperonin containing TCP1, subunit 5 (epsilon); K09497 T-complex
protein 1 subunit epsilon
Length=541
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 51/116 (43%), Gaps = 16/116 (13%)
Query 20 LTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPELIE 75
+ + + + +T +D E+R + F +K+ G VG + E V +DK+F +MP+ +E
Sbjct 185 MAEIAVNAVLTVADMERRDVDFELIKVEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVE 244
Query 76 FLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IL PPK + +H+ E + L Y K+ F ++
Sbjct 245 DAKIAILTCP------------FEPPKPKTKHKLDVTSVEDYKALQKYEKEKFEEM 288
> sce:YDR020C DAS2, RRT3; Das2p (EC:2.7.1.48)
Length=232
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 32 SDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDKEFLRREMPELIE 75
SDA++R+I+ +K VG+ E + + +MD LR EM + IE
Sbjct 122 SDADKRLISLIKKKNVGSNEQLAQLITEYMDH--LRPEMQQYIE 163
> tgo:TGME49_115860 EF hand domain-containing protein
Length=6006
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 32/70 (45%), Gaps = 7/70 (10%)
Query 66 LRREMP-ELIEF-----LHYRILDISSIKVVAKSW-FPRVAPPKKRYRHRALADIQESIE 118
LRRE+P + F LH D ++ VA SW FPR P + Y +DI +
Sbjct 988 LRREVPWSHVMFAWEKGLHLGEGDRLFLEHVAVSWAFPRQRPSVELYMSGEKSDIADLFP 1047
Query 119 ELAYYRKQIF 128
E Y R +F
Sbjct 1048 EFVYLRDLVF 1057
> dre:566080 myh11; myosin, heavy chain 11, smooth muscle
Length=1936
Score = 28.5 bits (62), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 108 RALADIQESIEELAYYRKQIFRDLEDSLTSRDN 140
RAL + Q S+ EL K + D+ED ++S+DN
Sbjct 1438 RALEECQGSLRELEKLNKTLRTDMEDLISSKDN 1470
> xla:100036778 cct5b; chaperonin containing TCP1, subunit 5 (epsilon)
b
Length=533
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 16/116 (13%)
Query 20 LTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPELIE 75
+ + + S +T +D +++ + F +K+ G VG K E V +DK+F +MP++++
Sbjct 185 MAEIAVNSILTVADMDRKDVDFELIKVEGKVGGKLEDTKLIKGVIVDKDFSHPQMPKVVK 244
Query 76 FLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IL PPK + +H+ E + L Y ++ F ++
Sbjct 245 DAKIAILTCP------------FEPPKPKTKHKLDVTSVEDYKALQKYEREKFLEM 288
> xla:380145 cct5, MGC53061, cct5a; chaperonin containing TCP1,
subunit 5 (epsilon); K09497 T-complex protein 1 subunit epsilon
Length=541
Score = 28.5 bits (62), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 16/116 (13%)
Query 20 LTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPELIE 75
+ + + S +T +D +++ + F +K+ G VG K E V +DK+F +MP++++
Sbjct 185 MAEIAVNSILTVADMDRKDVDFELIKVEGKVGGKLEDTKLIKGVIVDKDFSHPQMPKVVK 244
Query 76 FLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IL PPK + +H+ E + L Y ++ F ++
Sbjct 245 DAKIAILTCP------------FEPPKPKTKHKLDVTSVEDYKALQKYEREKFLEM 288
> cel:C06G3.6 hypothetical protein
Length=498
Score = 28.1 bits (61), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 2/31 (6%)
Query 57 NSVHMDKEFLRREMPELIEFLHYRILDISSI 87
N++ + F ++P+L FLH RILD+S I
Sbjct 84 NNIRPELSFYHSQLPQL--FLHVRILDVSVI 112
Lambda K H
0.320 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40