bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2191_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
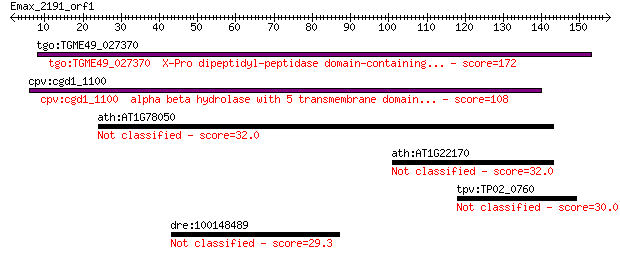
tgo:TGME49_027370 X-Pro dipeptidyl-peptidase domain-containing... 172 4e-43
cpv:cgd1_1100 alpha beta hydrolase with 5 transmembrane domain... 108 7e-24
ath:AT1G78050 PGM; PGM (PHOSPHOGLYCERATE/BISPHOSPHOGLYCERATE M... 32.0 0.81
ath:AT1G22170 phosphoglycerate/bisphosphoglycerate mutase fami... 32.0 0.94
tpv:TP02_0760 hypothetical protein 30.0 2.9
dre:100148489 nucleotide-binding oligomerization domain contai... 29.3 5.2
> tgo:TGME49_027370 X-Pro dipeptidyl-peptidase domain-containing
protein (EC:3.4.14.11); K06978
Length=1177
Score = 172 bits (436), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 74/146 (50%), Positives = 103/146 (70%), Gaps = 1/146 (0%)
Query 8 AQPSSFGITHNIMQKIAQKKVSVLAVGGFCDSATARGAIRLFSHIQENAPETRSQLILGN 67
A + FGI +M+K+A++ VSV AVGG+CDSA+ RGA RL+ +++ NAPE+RS+L+LG+
Sbjct 463 ASAAEFGIHEAVMRKLAERNVSVYAVGGYCDSASVRGASRLYEYMKRNAPESRSKLLLGS 522
Query 68 WSHGGRRSCDPFSGSFSCFEENLYKTILRFLDCQLKNKCWGGIQQEEPVHYWQTGTGEWR 127
WSHGGRRSC P++GSF CFE + Y +RF DC LK +CWG I E VHYWQ G+ EWR
Sbjct 523 WSHGGRRSCSPYAGSFPCFEADQYLDAVRFFDCHLKGQCWGNINDEPSVHYWQVGSEEWR 582
Query 128 NAYTFPPP-NVEHYDLRLSPQPLATM 152
+ +PP ++ + D LS + +
Sbjct 583 TSEYYPPARHMRYVDFHLSNEKFVNL 608
> cpv:cgd1_1100 alpha beta hydrolase with 5 transmembrane domains
; K06978
Length=1103
Score = 108 bits (270), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 53/151 (35%), Positives = 80/151 (52%), Gaps = 21/151 (13%)
Query 6 LYAQPSSFGITHNIMQKIAQKKVSVLAVGGFCDSATARGAIRLFSHIQENAPETRSQ--- 62
+YA+ G T ++ + K V+V G+CDSA +RG I+ + + E+A + Q
Sbjct 673 IYAE--RLGNTEDVATILGNKGVNVYLTSGYCDSANSRGVIQFYDALVESASKAYEQYLL 730
Query 63 --------------LILGNWSHGGRRSCDPFSGSFSCFEENLYKTILRFLDCQLKNKCWG 108
L+LG W+H GR SC PF S SCFE LY ++RF+DC LK CW
Sbjct 731 EAGNMASKEKPIYKLVLGPWTHSGRASCSPFGKSISCFEPALYYDLVRFMDCALKGICWE 790
Query 109 GIQQEEPVHYWQTGTGEWRNAYTFPPPNVEH 139
Q++ +H++Q G +W FPPP+ ++
Sbjct 791 --NQDKNIHFFQVGEEKWYTTDKFPPPDTKY 819
> ath:AT1G78050 PGM; PGM (PHOSPHOGLYCERATE/BISPHOSPHOGLYCERATE
MUTASE); catalytic/ intramolecular transferase, phosphotransferases
Length=332
Score = 32.0 bits (71), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 32/127 (25%), Positives = 50/127 (39%), Gaps = 19/127 (14%)
Query 24 AQKKVSVLAVGGFCDSATARG----AIRLFSHIQENAPETRSQLILGNWSHGGRRSCDPF 79
A KK+S + V S+ R + + H ++ P +IL N S + F
Sbjct 116 AGKKISNIPVDLIFTSSLIRAQMTAMLAMTQHRRKKVP-----IILHNESVKAKTWSHVF 170
Query 80 SGSFSCFEENLYKTILRFLDCQLKNKCWGGIQ----QEEPVHYWQTGTGEWRNAYTFPPP 135
S EE ++I QL + +G +Q +E Y EWR +Y PPP
Sbjct 171 S------EETRKQSIPVIAAWQLNERMYGELQGLNKKETAERYGTQQVHEWRRSYEIPPP 224
Query 136 NVEHYDL 142
E ++
Sbjct 225 KGESLEM 231
> ath:AT1G22170 phosphoglycerate/bisphosphoglycerate mutase family
protein
Length=334
Score = 32.0 bits (71), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 22/46 (47%), Gaps = 4/46 (8%)
Query 101 QLKNKCWGGIQ----QEEPVHYWQTGTGEWRNAYTFPPPNVEHYDL 142
QL + +G +Q QE Y + EWR +Y PPP E ++
Sbjct 185 QLNERMYGELQGLNKQETAERYGKEQVHEWRRSYDIPPPKGESLEM 230
> tpv:TP02_0760 hypothetical protein
Length=1331
Score = 30.0 bits (66), Expect = 2.9, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 118 YWQTGTGEWRNAYTFPPPNVEHYDLRLSPQP 148
Y++ GEW N+Y+F ++H L S QP
Sbjct 62 YFEKKNGEWNNSYSFTRKLLDHDQLLSSTQP 92
> dre:100148489 nucleotide-binding oligomerization domain containing
2-like
Length=991
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 21/44 (47%), Gaps = 3/44 (6%)
Query 43 RGAIRLFSHIQENAPETRSQLILGNWSHGGRRSCDPFSGSFSCF 86
+ ++L H E+ TR +L NW HGG P ++CF
Sbjct 779 QSGVQLLQHTLEDPHYTRQKL---NWDHGGHFRITPGLRKYACF 819
Lambda K H
0.319 0.134 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3516928200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40