bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2178_orf1
Length=67
Score E
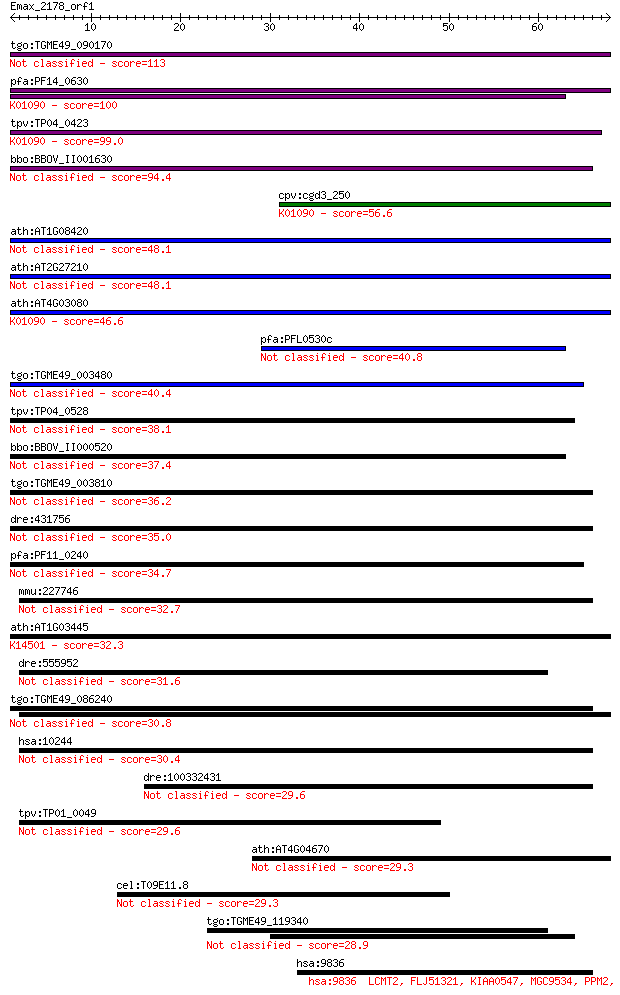
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090170 protein serine/threonine phosphatase, putati... 113 1e-25
pfa:PF14_0630 protein serine/threonine phosphatase; K01090 pro... 100 1e-21
tpv:TP04_0423 serine/threonine protein phosphatase (EC:3.1.3.1... 99.0 3e-21
bbo:BBOV_II001630 18.m09939; kelch repeat domain containing/Se... 94.4 8e-20
cpv:cgd3_250 protein serine/threonine phosphatase alpha ; K010... 56.6 2e-08
ath:AT1G08420 BSL2; kelch repeat-containing protein / serine/t... 48.1 7e-06
ath:AT2G27210 BSL3; kelch repeat-containing serine/threonine p... 48.1 7e-06
ath:AT4G03080 BSL1; kelch repeat-containing serine/threonine p... 46.6 2e-05
pfa:PFL0530c conserved Plasmodium protein 40.8 0.001
tgo:TGME49_003480 hypothetical protein 40.4 0.001
tpv:TP04_0528 hypothetical protein 38.1 0.007
bbo:BBOV_II000520 18.m06027; hypothetical protein 37.4 0.011
tgo:TGME49_003810 kelch motif domain-containing protein 36.2 0.030
dre:431756 rabepk; zgc:91813 35.0 0.065
pfa:PF11_0240 dynein heavy chain, putative 34.7 0.081
mmu:227746 Rabepk, 8430412M01Rik, 9530020D24Rik, AV073337, C87... 32.7 0.27
ath:AT1G03445 BSU1; BSU1 (BRI1 SUPPRESSOR 1); protein serine/t... 32.3 0.36
dre:555952 si:dkey-3d4.3 31.6 0.73
tgo:TGME49_086240 leucine-zipper-like transcriptional regulato... 30.8 1.1
hsa:10244 RABEPK, DKFZp686P1077, RAB9P40, bA65N13.1, p40; Rab9... 30.4 1.5
dre:100332431 multiple EGF-like-domains 8-like 29.6 2.4
tpv:TP01_0049 hypothetical protein 29.6 2.5
ath:AT4G04670 Met-10+ like family protein / kelch repeat-conta... 29.3 2.9
cel:T09E11.8 hypothetical protein 29.3 3.2
tgo:TGME49_119340 kelch motif domain-containing protein 28.9 4.1
hsa:9836 LCMT2, FLJ51321, KIAA0547, MGC9534, PPM2, TYW4; leuci... 27.7 9.4
> tgo:TGME49_090170 protein serine/threonine phosphatase, putative
/ sortilin (EC:3.1.3.16)
Length=931
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 54/67 (80%), Positives = 58/67 (86%), Gaps = 0/67 (0%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
VVFGGATGGGSLS EELYLL LRK+P LQW+ VPLQG TPGRRYGH M Y+ PNII+ GG
Sbjct 119 VVFGGATGGGSLSAEELYLLDLRKDPELQWMPVPLQGITPGRRYGHSMVYNKPNIIVFGG 178
Query 61 NDGERPL 67
NDGERPL
Sbjct 179 NDGERPL 185
> pfa:PF14_0630 protein serine/threonine phosphatase; K01090 protein
phosphatase [EC:3.1.3.16]
Length=889
Score = 100 bits (249), Expect = 1e-21, Method: Composition-based stats.
Identities = 43/67 (64%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
V++GGATGGGSLS ++LY+L LRKE W+ VP +G TPGRRYGHVM YS PN+I+ GG
Sbjct 93 VIYGGATGGGSLSLDDLYILDLRKEQKYTWMTVPTKGVTPGRRYGHVMVYSKPNLIVFGG 152
Query 61 NDGERPL 67
NDG+ L
Sbjct 153 NDGQNTL 159
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 30/63 (47%), Gaps = 2/63 (3%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQ-GSTPGRRYGHVMGYSTPNIILHG 59
V+FGG + S ++ + L ++ W+ P++ GS P RY H + I + G
Sbjct 207 VIFGGRSAENK-SLDDTWGLRQHRDGRWDWVEAPIKKGSPPEARYQHTSVFIGSKIFILG 265
Query 60 GND 62
G +
Sbjct 266 GRN 268
> tpv:TP04_0423 serine/threonine protein phosphatase (EC:3.1.3.16);
K01090 protein phosphatase [EC:3.1.3.16]
Length=798
Score = 99.0 bits (245), Expect = 3e-21, Method: Composition-based stats.
Identities = 42/66 (63%), Positives = 53/66 (80%), Gaps = 0/66 (0%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
VVFGGATGGG+LS ++L+LL LR+E L W+ VP G +PGRRYGH M +S PN+IL GG
Sbjct 90 VVFGGATGGGALSSDDLFLLDLRREKQLSWIIVPTTGRSPGRRYGHTMVFSKPNLILIGG 149
Query 61 NDGERP 66
NDG++P
Sbjct 150 NDGQQP 155
> bbo:BBOV_II001630 18.m09939; kelch repeat domain containing/Serine/threonine
protein phosphatase protein
Length=799
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 41/65 (63%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
VVFGGATGGGSLS ++LYLL LR++ HL W+ VP G +PGRRYGH M +S PN+++ GG
Sbjct 85 VVFGGATGGGSLSSDDLYLLDLRRDKHLSWITVPTTGRSPGRRYGHTMVFSKPNLVVIGG 144
Query 61 NDGER 65
NDG++
Sbjct 145 NDGQQ 149
> cpv:cgd3_250 protein serine/threonine phosphatase alpha ; K01090
protein phosphatase [EC:3.1.3.16]
Length=772
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 24/38 (63%), Positives = 31/38 (81%), Gaps = 1/38 (2%)
Query 31 LAVPL-QGSTPGRRYGHVMGYSTPNIILHGGNDGERPL 67
++VP+ G TPGRRYGH+M YS PN+I+ GGNDG+R L
Sbjct 1 MSVPICGGRTPGRRYGHIMVYSKPNLIVFGGNDGQRTL 38
> ath:AT1G08420 BSL2; kelch repeat-containing protein / serine/threonine
phosphoesterase family protein
Length=1018
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/68 (44%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILH-G 59
VV G G LS E+L++L L ++ +W V +QG PG RYGHVM ++ G
Sbjct 200 VVIQGGIGPAGLSAEDLHVLDLTQQ-RPRWHRVVVQGPGPGPRYGHVMALVGQRYLMAIG 258
Query 60 GNDGERPL 67
GNDG+RPL
Sbjct 259 GNDGKRPL 266
> ath:AT2G27210 BSL3; kelch repeat-containing serine/threonine
phosphoesterase family protein
Length=1006
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/68 (44%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILH-G 59
VV G G LS E+L++L L ++ +W V +QG PG RYGHVM ++ G
Sbjct 189 VVIQGGIGPAGLSAEDLHVLDLTQQ-RPRWHRVVVQGPGPGPRYGHVMALVGQRYLMAIG 247
Query 60 GNDGERPL 67
GNDG+RPL
Sbjct 248 GNDGKRPL 255
> ath:AT4G03080 BSL1; kelch repeat-containing serine/threonine
phosphoesterase family protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=881
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMG-YSTPNIILHG 59
VVF G G S ++LY+L + + +W V +QG PG RYGHVM S ++
Sbjct 111 VVFQGGIGPAGHSTDDLYVLDMTND-KFKWHRVVVQGDGPGPRYGHVMDLVSQRYLVTVT 169
Query 60 GNDGERPL 67
GNDG+R L
Sbjct 170 GNDGKRAL 177
> pfa:PFL0530c conserved Plasmodium protein
Length=1065
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 29 QWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGND 62
+W+ + ++G P +RYGH + + P+++L GGN+
Sbjct 399 KWVEIKIKGKKPSKRYGHSLDFLYPHLVLFGGNE 432
> tgo:TGME49_003480 hypothetical protein
Length=1325
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 32/64 (50%), Gaps = 10/64 (15%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
+FGG GG L+ + L+W A G P RYGH + Y+ P++IL GG
Sbjct 572 ALFGGKREGGELADNQ----------ELRWAAPAFVGPLPSPRYGHSLVYAAPHLILFGG 621
Query 61 NDGE 64
DG+
Sbjct 622 VDGD 625
> tpv:TP04_0528 hypothetical protein
Length=753
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 31/63 (49%), Gaps = 1/63 (1%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
+V GG T L L++L L EP L W G +P +R+GH M +++ GG
Sbjct 337 LVCGGFTHNKLLLDTHLHVLNLTTEP-LTWTIFQTSGPSPPKRFGHSMAQVGNYVVIFGG 395
Query 61 NDG 63
+G
Sbjct 396 CNG 398
> bbo:BBOV_II000520 18.m06027; hypothetical protein
Length=310
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTP-GRRYGHVMGYSTPNIILHG 59
+++GG T S LY LG+ L+W +P+ P RR H M Y I++ G
Sbjct 74 IIYGGITDDDS---SRLYALGMDNHGELRWAILPITAKGPIERRAFHSMTYGNGEIVITG 130
Query 60 GND 62
G D
Sbjct 131 GED 133
> tgo:TGME49_003810 kelch motif domain-containing protein
Length=531
Score = 36.2 bits (82), Expect = 0.030, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 30/65 (46%), Gaps = 6/65 (9%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
++FGG G LS Y P W AV + P R +GHV + + +HGG
Sbjct 208 ILFGGCDDGEQLSTTYKY-----DPPMRTWTAVQTEHKPPAR-HGHVALVNDGRVYIHGG 261
Query 61 NDGER 65
DG+R
Sbjct 262 YDGDR 266
> dre:431756 rabepk; zgc:91813
Length=351
Score = 35.0 bits (79), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 32/65 (49%), Gaps = 3/65 (4%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
V GG G +++ +L++ + W G+ P +R+GHV+ +I +HGG
Sbjct 164 VFSGGEAGSSAVTDAQLHVFDAVS---VTWTQPDTTGTPPAQRHGHVITAVGSDIYIHGG 220
Query 61 NDGER 65
GE+
Sbjct 221 MSGEK 225
> pfa:PF11_0240 dynein heavy chain, putative
Length=5251
Score = 34.7 bits (78), Expect = 0.081, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
++FGG G S +LY + +W+ +G+ P RYGH+ N+ + GG
Sbjct 268 ILFGGYGGYDRSSYNDLYEFNILNN---EWILRGCKGNVPSTRYGHISFIHGNNLFILGG 324
Query 61 NDGE 64
+ E
Sbjct 325 TNAE 328
> mmu:227746 Rabepk, 8430412M01Rik, 9530020D24Rik, AV073337, C87311,
Rab9p40; Rab9 effector protein with kelch motifs
Length=380
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 2 VFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGN 61
VFGG G P E L + L W GS P R+GHVM + + +HGG
Sbjct 162 VFGGGERGAQ--PVEDVKLHVFDANTLTWSQPETHGSPPSPRHGHVMVAAGTKLFIHGGL 219
Query 62 DGER 65
G++
Sbjct 220 AGDK 223
> ath:AT1G03445 BSU1; BSU1 (BRI1 SUPPRESSOR 1); protein serine/threonine
phosphatase; K14501 serine/threonine-protein phosphatase
BSU1 [EC:3.1.3.16]
Length=793
Score = 32.3 bits (72), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 33/68 (48%), Gaps = 4/68 (5%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPN-IILHG 59
++ G G S ++Y+L + +W+ + G TP RYGHVM + +++
Sbjct 111 ILIQGGIGPSGPSDGDVYMLDMTNN---KWIKFLVGGETPSPRYGHVMDIAAQRWLVIFS 167
Query 60 GNDGERPL 67
GN+G L
Sbjct 168 GNNGNEIL 175
> dre:555952 si:dkey-3d4.3
Length=422
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Query 2 VFGGATGGG-SLSPEELYLLGLRKEPHLQWLA--VPLQGSTPGRRYGHVMGYSTPNIILH 58
VFGG S S L++ KE + W A V LQG TPG R GH ++ ++
Sbjct 87 VFGGLRDSSYSNSKTPLWVFDTGKECWINWKAASVALQGVTPGNRKGHSAVILGSSMYIY 146
Query 59 GG 60
GG
Sbjct 147 GG 148
> tgo:TGME49_086240 leucine-zipper-like transcriptional regulator
1, putative
Length=625
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 6/67 (8%)
Query 2 VFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQG-STPGRRYGHVMGYSTPNIILHGG 60
+FGG G +L+ +L++L ++ L+W V G S P R H I +HGG
Sbjct 122 IFGGWNGKFALN--DLFILDVQS---LRWTRVEQDGCSPPEARNNHTTAAVGDRIFVHGG 176
Query 61 NDGERPL 67
+DG + L
Sbjct 177 HDGTQWL 183
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 27/65 (41%), Gaps = 5/65 (7%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
V+FGG +G L+ ++ L W + GS P GH I L GG
Sbjct 280 VLFGGHSGNTHLTDLHVF-----DTATLTWTKPEISGSPPPGLRGHTANLIGHKIFLFGG 334
Query 61 NDGER 65
DG+R
Sbjct 335 YDGKR 339
> hsa:10244 RABEPK, DKFZp686P1077, RAB9P40, bA65N13.1, p40; Rab9
effector protein with kelch motifs
Length=372
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 2 VFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGN 61
VFGG G P + L + L W G+ P R+GHVM + + +HGG
Sbjct 154 VFGGGERGAQ--PVQDTKLHVFDANTLTWSQPETLGNPPSPRHGHVMVAAGTKLFIHGGL 211
Query 62 DGER 65
G+R
Sbjct 212 AGDR 215
> dre:100332431 multiple EGF-like-domains 8-like
Length=2834
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 25/51 (49%), Gaps = 4/51 (7%)
Query 16 ELYLLGLRKEPHLQW-LAVPLQGSTPGRRYGHVMGYSTPNIILHGGNDGER 65
ELY L P+L W L VP QG+ P R+ H +++ GG GE
Sbjct 1686 ELYSLYY---PNLTWSLLVPSQGNKPLSRFFHAAALVKDTMVIVGGRTGEE 1733
> tpv:TP01_0049 hypothetical protein
Length=134
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 3/47 (6%)
Query 2 VFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVM 48
+FGG G L++L +P+L+W + GS P RR G +
Sbjct 52 MFGGEIDGEGRYTNNLFVLN---DPNLEWSILSANGSFPSRRSGSTL 95
> ath:AT4G04670 Met-10+ like family protein / kelch repeat-containing
protein
Length=995
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 18/40 (45%), Gaps = 0/40 (0%)
Query 28 LQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGNDGERPL 67
LQW V QG P R+ H M + GG +GE L
Sbjct 411 LQWKEVEQQGQWPCARHSHAMVAYGSQSFMFGGYNGENVL 450
> cel:T09E11.8 hypothetical protein
Length=543
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 13 SPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMG 49
SP +LY+ L E + ++ VP++ PG Y H +G
Sbjct 144 SPHQLYI-NLSTEGNNEYTVVPVKDVRPGPTYSHTLG 179
> tgo:TGME49_119340 kelch motif domain-containing protein
Length=643
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 18/38 (47%), Gaps = 0/38 (0%)
Query 23 RKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGG 60
R+EP + W G+ P RYGH +I+ GG
Sbjct 419 RREPDMTWSQPETSGTPPCPRYGHASLVVGRQLIVCGG 456
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 30 WLAVPLQGSTPGRRYGHVMGYSTPNIILHGGNDG 63
W V G+ P R+GH M ++++ GG G
Sbjct 494 WYRVRTHGNPPAPRFGHTMAALNGDLVVFGGWAG 527
> hsa:9836 LCMT2, FLJ51321, KIAA0547, MGC9534, PPM2, TYW4; leucine
carboxyl methyltransferase 2; K00599 [EC:2.1.1.-]
Length=686
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Query 33 VPLQGSTPG-RRYGHVMGYSTPNIILHGGNDGER 65
V +G P +RYGH + +P++IL G GE+
Sbjct 346 VSSEGQVPNLKRYGHASVFLSPDVILSAGGFGEQ 379
Lambda K H
0.317 0.144 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2038271796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40