bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2159_orf1
Length=112
Score E
Sequences producing significant alignments: (Bits) Value
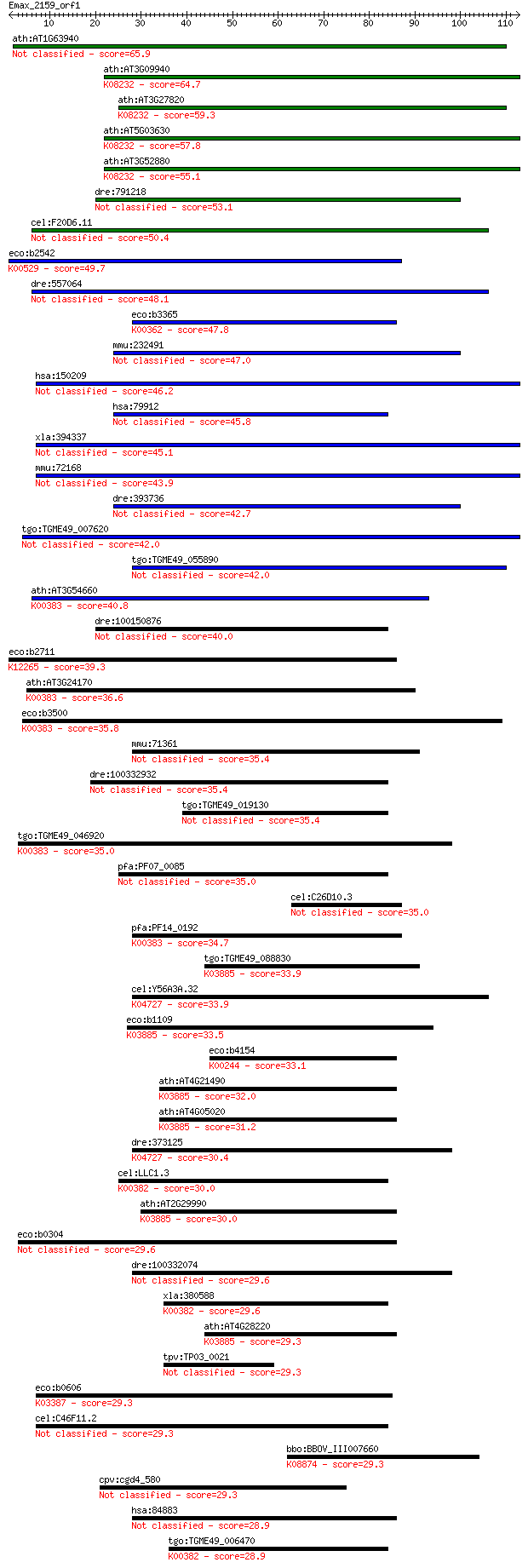
ath:AT1G63940 monodehydroascorbate reductase, putative 65.9 3e-11
ath:AT3G09940 MDHAR; MDHAR (MONODEHYDROASCORBATE REDUCTASE); m... 64.7 8e-11
ath:AT3G27820 MDAR4; MDAR4 (MONODEHYDROASCORBATE REDUCTASE 4);... 59.3 3e-09
ath:AT5G03630 ATMDAR2; monodehydroascorbate reductase (NADH); ... 57.8 8e-09
ath:AT3G52880 ATMDAR1; monodehydroascorbate reductase, putativ... 55.1 6e-08
dre:791218 zgc:158614 53.1 2e-07
cel:F20D6.11 hypothetical protein 50.4 1e-06
eco:b2542 hcaD, ECK2539, hcaA4, JW2526, phdA, yfhY; phenylprop... 49.7 3e-06
dre:557064 apoptosis-inducing factor, mitochondrion-associated... 48.1 7e-06
eco:b3365 nirB, ECK3353, JW3328; nitrite reductase, large subu... 47.8 9e-06
mmu:232491 Pyroxd1; pyridine nucleotide-disulphide oxidoreduct... 47.0 2e-05
hsa:150209 AIFM3, AIFL, FLJ30473, FLJ45137; apoptosis-inducing... 46.2 3e-05
hsa:79912 PYROXD1, FLJ22028; pyridine nucleotide-disulphide ox... 45.8 3e-05
xla:394337 aifm3, MGC84340, nfrl-A; apoptosis-inducing factor,... 45.1 5e-05
mmu:72168 Aifm3, 2810401C16Rik, AI840249, Aifl; apoptosis-indu... 43.9 1e-04
dre:393736 MGC73254, pyroxd1; zgc:73254 (EC:1.8.1.-) 42.7 3e-04
tgo:TGME49_007620 pyridine nucleotide-disulphide oxidoreductas... 42.0 5e-04
tgo:TGME49_055890 monodehydroascorbate reductase, putative (EC... 42.0 5e-04
ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding / gl... 40.8 0.001
dre:100150876 apoptosis-inducing factor, mitochondrion-associa... 40.0 0.002
eco:b2711 norW, ECK2706, JW2681, ygaL, ygbD; NADH:flavorubredo... 39.3 0.003
ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase); ... 36.6 0.022
eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreducta... 35.8 0.032
mmu:71361 Aifm2, 5430437E11Rik, Amid, D730001I10Rik, PRG3; apo... 35.4 0.040
dre:100332932 glutathione reductase-like 35.4 0.049
tgo:TGME49_019130 glutathione reductase, putative (EC:1.8.1.7) 35.4 0.051
tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7)... 35.0 0.055
pfa:PF07_0085 ferrodoxin reductase-like protein 35.0 0.059
cel:C26D10.3 hypothetical protein 35.0 0.063
pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 gluta... 34.7 0.078
tgo:TGME49_088830 pyridine nucleotide-disulphide oxidoreductas... 33.9 0.12
cel:Y56A3A.32 wah-1; Worm AIF (apoptosis inducing factor) Homo... 33.9 0.14
eco:b1109 ndh, ECK1095, JW1095; respiratory NADH dehydrogenase... 33.5 0.18
eco:b4154 frdA, ECK4150, JW4115; fumarate reductase (anaerobic... 33.1 0.23
ath:AT4G21490 NDB3; NDB3; NADH dehydrogenase; K03885 NADH dehy... 32.0 0.51
ath:AT4G05020 NDB2; NDB2 (NAD(P)H dehydrogenase B2); FAD bindi... 31.2 0.79
dre:373125 pdcd8, AIF, zgc:91994; programmed cell death 8 (apo... 30.4 1.6
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 30.0 1.7
ath:AT2G29990 NDA2; NDA2 (ALTERNATIVE NAD(P)H DEHYDROGENASE 2)... 30.0 1.9
eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide... 29.6 2.2
dre:100332074 programmed cell death 8 (apoptosis-inducing fact... 29.6 2.2
xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1... 29.6 2.6
ath:AT4G28220 NDB1; NDB1 (NAD(P)H dehydrogenase B1); NADH dehy... 29.3 2.9
tpv:TP03_0021 hypothetical protein 29.3 2.9
eco:b0606 ahpF, ECK0600, JW0599; alkyl hydroperoxide reductase... 29.3 3.1
cel:C46F11.2 hypothetical protein 29.3 3.1
bbo:BBOV_III007660 17.m07668; hypothetical protein; K08874 tra... 29.3 3.2
cpv:cgd4_580 hypothetical protein 29.3 3.3
hsa:84883 AIFM2, AMID, DKFZp686L1298, PRG3, RP11-367H5.2; apop... 28.9 3.9
tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.... 28.9 4.1
> ath:AT1G63940 monodehydroascorbate reductase, putative
Length=486
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 45/111 (40%), Positives = 64/111 (57%), Gaps = 5/111 (4%)
Query 2 HRAHGVEIHLNAKMDCL-IGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPL-IAAGA 59
+R +GV+ A ++ L G DG V+ V+++DGS + AD VV+GIG PA+ P A
Sbjct 261 YRQNGVKFVKGASINNLEAGSDGRVSAVKLADGSTIEADTVVIGIGAKPAIGPFETLAMN 320
Query 60 ASGNGVEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRI-RLESVQNANDQA 109
S G++VD +TS P I+AIGD A A P R+ R+E V +A A
Sbjct 321 KSIGGIQVDGLFRTSTPGIFAIGDVA--AFPLKIYDRMTRVEHVDHARRSA 369
> ath:AT3G09940 MDHAR; MDHAR (MONODEHYDROASCORBATE REDUCTASE);
monodehydroascorbate reductase (NADH); K08232 monodehydroascorbate
reductase (NADH) [EC:1.6.5.4]
Length=433
Score = 64.7 bits (156), Expect = 8e-11, Method: Composition-based stats.
Identities = 35/91 (38%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Query 22 DGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVDEFCQTSLPNIYAI 81
DG VT V++ DG L A++VV G+G PA G++ D F +TS+P++YA+
Sbjct 230 DGEVTEVKLEDGRTLEANIVVAGVGARPATSLFKGQLEEEKGGIKTDGFFKTSVPDVYAL 289
Query 82 GDCAIHANPFAGGQRIRLESVQNANDQAITA 112
GD A GG R R+E NA A A
Sbjct 290 GDVATFPMKMYGGTR-RVEHADNARKSAAQA 319
> ath:AT3G27820 MDAR4; MDAR4 (MONODEHYDROASCORBATE REDUCTASE 4);
monodehydroascorbate reductase (NADH); K08232 monodehydroascorbate
reductase (NADH) [EC:1.6.5.4]
Length=488
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 36/85 (42%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query 25 VTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVDEFCQTSLPNIYAIGDC 84
VT V + DGS LPAD+VVVGIGI P G++V+ Q+S ++YAIGD
Sbjct 238 VTAVNLKDGSHLPADLVVVGIGIRPNTSLFEGQLTIEKGGIKVNSRMQSSDSSVYAIGDV 297
Query 85 AIHANPFAGGQRIRLESVQNANDQA 109
A G R RLE V +A A
Sbjct 298 ATFPVKLFGEMR-RLEHVDSARKSA 321
> ath:AT5G03630 ATMDAR2; monodehydroascorbate reductase (NADH);
K08232 monodehydroascorbate reductase (NADH) [EC:1.6.5.4]
Length=435
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 35/94 (37%), Positives = 51/94 (54%), Gaps = 4/94 (4%)
Query 22 DGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVDEFCQTSLPNIYAI 81
+G VT V++ DG L AD+V+VG+G P + G++ D F +TSLP++YAI
Sbjct 237 NGEVTEVKLKDGRTLEADIVIVGVGGRPIISLFKDQVEEEKGGLKTDGFFKTSLPDVYAI 296
Query 82 GDCAIHANPFAGGQRIRLESVQNAN---DQAITA 112
GD A R R+E V +A +QA+ A
Sbjct 297 GDVATFPMKLYNEMR-RVEHVDHARKSAEQAVKA 329
> ath:AT3G52880 ATMDAR1; monodehydroascorbate reductase, putative;
K08232 monodehydroascorbate reductase (NADH) [EC:1.6.5.4]
Length=466
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 50/94 (53%), Gaps = 4/94 (4%)
Query 22 DGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVDEFCQTSLPNIYAI 81
+G V V++ DG L AD+V+VG+G P G++ D F +TS+P++YA+
Sbjct 268 NGEVKEVQLKDGRTLEADIVIVGVGAKPLTSLFKGQVEEDKGGIKTDAFFKTSVPDVYAV 327
Query 82 GDCAIHANPFAGGQRIRLESVQNAN---DQAITA 112
GD A G R R+E V ++ +QA+ A
Sbjct 328 GDVATFPLKMYGDVR-RVEHVDHSRKSAEQAVKA 360
> dre:791218 zgc:158614
Length=455
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 46/82 (56%), Gaps = 2/82 (2%)
Query 20 GHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPL--IAAGAASGNGVEVDEFCQTSLPN 77
G +G V V + +G VLPAD+++ GIG++P + L S V VD+F +T++P+
Sbjct 261 GENGKVKEVVLKNGEVLPADIIIAGIGVIPNSDFLKETLVEIDSRKAVVVDKFMKTNIPD 320
Query 78 IYAIGDCAIHANPFAGGQRIRL 99
++A GD G +R+ +
Sbjct 321 VFAAGDVVSFPLTLVGHKRVNI 342
> cel:F20D6.11 hypothetical protein
Length=549
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 54/103 (52%), Gaps = 3/103 (2%)
Query 6 GVEIHLNAKMDCLIGHD-GWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNG 64
GV+ L A + L G+D G V+ V + +G L D++V GIG+ PA + L +G N
Sbjct 337 GVKFELAANVVALRGNDQGEVSKVILENGKELDVDLLVCGIGVTPATKFLEGSGIKLDNR 396
Query 65 --VEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQNA 105
+EVDE +T++ I+A+GD P I ++ Q A
Sbjct 397 GFIEVDEKFRTNISYIFAMGDVVTAPLPLWDIDSINIQHFQTA 439
> eco:b2542 hcaD, ECK2539, hcaA4, JW2526, phdA, yfhY; phenylpropionate
dioxygenase, ferredoxin reductase subunit (EC:1.18.1.3);
K00529 ferredoxin--NAD+ reductase [EC:1.18.1.3]
Length=400
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 46/86 (53%), Gaps = 2/86 (2%)
Query 1 EHRAHGVEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAA 60
H+ GV I LN ++ ++ DG + + G L AD+V+ GIGI + A
Sbjct 195 RHQQAGVRILLNNAIEHVV--DGEKVELTLQSGETLQADVVIYGIGISANEQLAREANLD 252
Query 61 SGNGVEVDEFCQTSLPNIYAIGDCAI 86
+ NG+ +DE C+T P I+A GD AI
Sbjct 253 TANGIVIDEACRTCDPAIFAGGDVAI 278
> dre:557064 apoptosis-inducing factor, mitochondrion-associated,
3-like
Length=573
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 52/102 (50%), Gaps = 2/102 (1%)
Query 6 GVEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAA--SGN 63
GV ++N + + G + V V++ G + AD+++V IG+ P E L + S N
Sbjct 363 GVTFYMNDAVAEVQGKNRRVKAVKLKSGITIEADLLIVAIGVSPNSEFLKGSRVRMDSKN 422
Query 64 GVEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQNA 105
V VDE+ +T++ ++Y GD A GQ++ L Q A
Sbjct 423 YVIVDEYMRTNITDVYCAGDLTSFPLKMAKGQKVSLGHWQIA 464
> eco:b3365 nirB, ECK3353, JW3328; nitrite reductase, large subunit,
NAD(P)H-binding (EC:1.7.1.4); K00362 nitrite reductase
(NAD(P)H) large subunit [EC:1.7.1.4]
Length=847
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query 28 VRMSDGSVLPADMVVVGIGIVPAVEPLIAAG--AASGNGVEVDEFCQTSLPNIYAIGDCA 85
+R +DGS L D +V GI P + G A G+ +++ CQTS P+IYAIG+CA
Sbjct 224 MRFADGSELEVDFIVFSTGIRPRDKLATQCGLDVAPRGGIVINDSCQTSDPDIYAIGECA 283
> mmu:232491 Pyroxd1; pyridine nucleotide-disulphide oxidoreductase
domain 1 (EC:1.8.1.-)
Length=498
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 40/80 (50%), Gaps = 4/80 (5%)
Query 24 WVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAA---GAASGNGVEVDEFCQTSLPNIYA 80
W V +++G++ D +V G+ P V P + G+ VD+ +TSLP+IYA
Sbjct 294 WPVYVELTNGTIYGCDFLVSATGVTPNVHPFLHRNNFALGEDGGLRVDDQMRTSLPDIYA 353
Query 81 IGD-CAIHANPFAGGQRIRL 99
GD C P Q++RL
Sbjct 354 AGDICTACWQPSPVWQQMRL 373
> hsa:150209 AIFM3, AIFL, FLJ30473, FLJ45137; apoptosis-inducing
factor, mitochondrion-associated, 3
Length=598
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 30/108 (27%), Positives = 54/108 (50%), Gaps = 2/108 (1%)
Query 7 VEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAA--SGNG 64
V+ ++ ++ L G +G + V + V+ AD+ VVGIG VPA L +G S
Sbjct 389 VKFYMQTEVSELRGQEGKLKEVVLKSSKVVRADVCVVGIGAVPATGFLRQSGIGLDSRGF 448
Query 65 VEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQNANDQAITA 112
+ V++ QT++P ++A GD + +++ + Q A+ Q A
Sbjct 449 IPVNKMMQTNVPGVFAAGDAVTFPLAWRNNRKVNIPHWQMAHAQGRVA 496
> hsa:79912 PYROXD1, FLJ22028; pyridine nucleotide-disulphide
oxidoreductase domain 1 (EC:1.8.1.-)
Length=500
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 3/63 (4%)
Query 24 WVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGA---ASGNGVEVDEFCQTSLPNIYA 80
W V +++ + D +V G+ P VEP + + G++VD+ TSLP+IYA
Sbjct 296 WPVYVELTNEKIYGCDFIVSATGVTPNVEPFLHGNSFDLGEDGGLKVDDHMHTSLPDIYA 355
Query 81 IGD 83
GD
Sbjct 356 AGD 358
> xla:394337 aifm3, MGC84340, nfrl-A; apoptosis-inducing factor,
mitochondrion-associated, 3
Length=605
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 30/108 (27%), Positives = 53/108 (49%), Gaps = 2/108 (1%)
Query 7 VEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAA--SGNG 64
V+ ++ ++ L +G + V + G VL AD+ V+GIG P L +G A S
Sbjct 389 VKFYMQTEVSELREQEGKLKEVVLKSGKVLRADVCVIGIGASPTTGFLKQSGVALDSRGY 448
Query 65 VEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQNANDQAITA 112
+ V++ QT++P ++A GD F +++ + Q A+ Q A
Sbjct 449 IPVNKMMQTNIPGVFAAGDVVTFPLAFRNNKKMNVPHWQMAHMQGRIA 496
> mmu:72168 Aifm3, 2810401C16Rik, AI840249, Aifl; apoptosis-inducing
factor, mitochondrion-associated 3
Length=598
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/108 (26%), Positives = 53/108 (49%), Gaps = 2/108 (1%)
Query 7 VEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAA--SGNG 64
V+ ++ ++ L +G + V + VL AD+ V+GIG VPA L +G S
Sbjct 389 VKFYMQTEVSELRAQEGKLQEVVLKSSKVLRADVCVLGIGAVPATGFLRQSGIGLDSRGF 448
Query 65 VEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQNANDQAITA 112
+ V++ QT++P ++A GD + +++ + Q A+ Q A
Sbjct 449 IPVNKMMQTNVPGVFAAGDAVTFPLAWRNNRKVNIPHWQMAHAQGRVA 496
> dre:393736 MGC73254, pyroxd1; zgc:73254 (EC:1.8.1.-)
Length=490
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Query 24 WVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAG---AASGNGVEVDEFCQTSLPNIYA 80
W V++++G + D +V G+VP +P + A+ G+ VD+ +TS +++A
Sbjct 286 WPAYVQLTNGKIYGCDFIVSATGVVPNTDPFLPGNNFDVAADLGLLVDDHMRTSEADVFA 345
Query 81 IGD-CAIHANPFAGGQRIRL 99
GD C+ P + Q++RL
Sbjct 346 AGDVCSAGWEPSSIWQQMRL 365
> tgo:TGME49_007620 pyridine nucleotide-disulphide oxidoreductase
domain-containing protein (EC:1.7.1.4 1.18.1.3)
Length=664
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 34/114 (29%), Positives = 55/114 (48%), Gaps = 7/114 (6%)
Query 4 AHGVEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGA---A 60
+ GV + +K+ VTGV ++ G ++ AD+V+VGIG VPA + L A
Sbjct 436 SKGVRFYPQSKVVGFTSSRDRVTGVELASGEIIQADVVIVGIGSVPATKFLADQSEFALA 495
Query 61 SGNGVEVDEFCQ-TSLPNIYAIGDCAIHANPFAG-GQRIRLESVQNANDQAITA 112
+ D + + P+++ GD A A P+ G++IR+E A Q A
Sbjct 496 RDGAIVTDPLLRLPANPDVFVAGDIA--AYPYVKTGEQIRVEHWAVAMQQGRVA 547
> tgo:TGME49_055890 monodehydroascorbate reductase, putative (EC:1.6.5.4)
Length=718
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 45/103 (43%), Gaps = 21/103 (20%)
Query 28 VRMSDGS--VLPADMVVVGIGIVPAVE----PLIAAGAASGNGVEVDE------------ 69
V +SDG+ VL AD VV +G P V+ L+ A G G+ VD+
Sbjct 431 VTLSDGTSHVLQADFVVAAVGSRPVVDFLNWQLMLADECVGGGILVDQNLQAFPSRDVQI 490
Query 70 ---FCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQNANDQA 109
Q P ++A+GD A GG+ +R E V NA A
Sbjct 491 AAASAQKPHPEVFALGDVAAFPQTRNGGRPVRYEHVWNARSMA 533
> ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding /
glutathione-disulfide reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=565
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 6/93 (6%)
Query 6 GVEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPL----IAAGAAS 61
G+E H + +I ++ S G+V V+ G P + L + A
Sbjct 315 GIEFHTEESPEAIIKAGDGSFSLKTSKGTVEGFSHVMFATGRKPNTKNLGLENVGVKMAK 374
Query 62 GNGVEVDEFCQTSLPNIYAIGDCA--IHANPFA 92
+EVDE+ QTS+P+I+A+GD I+ P A
Sbjct 375 NGAIEVDEYSQTSVPSIWAVGDVTDRINLTPVA 407
> dre:100150876 apoptosis-inducing factor, mitochondrion-associated,
3
Length=599
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 2/66 (3%)
Query 20 GHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAA--SGNGVEVDEFCQTSLPN 77
GH+G + V + G VL AD+ V+GIG PA L +G S + V++ QT++
Sbjct 402 GHNGQLKEVVLKSGKVLRADVCVIGIGSSPATAFLKQSGVHIDSKGFIPVNKTMQTNIDG 461
Query 78 IYAIGD 83
++A GD
Sbjct 462 VFAGGD 467
> eco:b2711 norW, ECK2706, JW2681, ygaL, ygbD; NADH:flavorubredoxin
oxidoreductase (EC:1.18.1.-); K12265 nitric oxide reductase
FlRd-NAD(+) reductase [EC:1.18.1.-]
Length=377
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 28/87 (32%), Positives = 42/87 (48%), Gaps = 3/87 (3%)
Query 1 EHRAHGVEIHLNAKMDCLIGHDGWVTGVR--MSDGSVLPADMVVVGIGIVPAVEPLIAAG 58
+HR + +HL K L G + +G++ + + D V+ G+ P AG
Sbjct 190 QHRLTEMGVHLLLKSQ-LQGLEKTDSGIQATLDRQRNIEVDAVIAATGLRPETALARRAG 248
Query 59 AASGNGVEVDEFCQTSLPNIYAIGDCA 85
GV VD + QTS +IYA+GDCA
Sbjct 249 LTINRGVCVDSYLQTSNTDIYALGDCA 275
> ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase);
FAD binding / NADP or NADPH binding / glutathione-disulfide
reductase/ oxidoreductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=499
Score = 36.6 bits (83), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 40/89 (44%), Gaps = 5/89 (5%)
Query 5 HGVEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPL----IAAGAA 60
GV +H + L D + V S G AD+V+ G P + L +
Sbjct 257 RGVNLHPQTSLTQLTKTDQGIK-VISSHGEEFVADVVLFATGRSPNTKRLNLEAVGVELD 315
Query 61 SGNGVEVDEFCQTSLPNIYAIGDCAIHAN 89
V+VDE+ +T++P+I+A+GD N
Sbjct 316 QAGAVKVDEYSRTNIPSIWAVGDATNRIN 344
> eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreductase
(EC:1.8.1.7); K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=450
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 56/114 (49%), Gaps = 10/114 (8%)
Query 4 AHGVEIHLNAKMDCLIGH-DGWVTGVRMSDGSVLPADMVVVGIGIVPAVEP--LIAAGAA 60
A G ++H NA ++ + DG +T + + DG D ++ IG PA + L AAG
Sbjct 220 AEGPQLHTNAIPKAVVKNTDGSLT-LELEDGRSETVDCLIWAIGREPANDNINLEAAGVK 278
Query 61 SGNG--VEVDEFCQTSLPNIYAIGDC--AIHANP--FAGGQRIRLESVQNANDQ 108
+ + VD++ T++ IYA+GD A+ P A G+R+ N D+
Sbjct 279 TNEKGYIVVDKYQNTNIEGIYAVGDNTGAVELTPVAVAAGRRLSERLFNNKPDE 332
> mmu:71361 Aifm2, 5430437E11Rik, Amid, D730001I10Rik, PRG3; apoptosis-inducing
factor, mitochondrion-associated 2
Length=373
Score = 35.4 bits (80), Expect = 0.040, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 32/67 (47%), Gaps = 4/67 (5%)
Query 28 VRMSDGSVLPADMVVVGIGI---VPAVEPLIAAGAASGNGVEVDEFCQTS-LPNIYAIGD 83
V G+ + +MV+V GI A + AS ++V+EF Q NIYAIGD
Sbjct 226 VETDKGTEVATNMVIVCNGIKINSSAYRSAFESRLASNGALKVNEFLQVEGYSNIYAIGD 285
Query 84 CAIHANP 90
CA P
Sbjct 286 CADTKEP 292
> dre:100332932 glutathione reductase-like
Length=461
Score = 35.4 bits (80), Expect = 0.049, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 36/69 (52%), Gaps = 4/69 (5%)
Query 19 IGHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPL--IAAGAASG--NGVEVDEFCQTS 74
+ DG +G L D V++ +G P L AAG A+ + VDE+ +TS
Sbjct 237 VSRDGEGLVAETKEGETLRVDTVMLALGRDPHTRGLGLEAAGVATDAHGAIIVDEYSRTS 296
Query 75 LPNIYAIGD 83
+P+I+A+GD
Sbjct 297 VPHIFALGD 305
> tgo:TGME49_019130 glutathione reductase, putative (EC:1.8.1.7)
Length=505
Score = 35.4 bits (80), Expect = 0.051, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 6/51 (11%)
Query 39 DMVVVGIGIVPAVEPL------IAAGAASGNGVEVDEFCQTSLPNIYAIGD 83
D V++ + PA+E L + +G ++VD F TS+P IYA+GD
Sbjct 276 DHVIMAVNPAPAIEDLGLEEAGVDIDVNNGGFIKVDAFQNTSIPGIYAVGD 326
> tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 35.0 bits (79), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 48/104 (46%), Gaps = 9/104 (8%)
Query 3 RAHGVEIHLNAKMDCLIGHDGWVTGVRMSDG-SVLPADMVVVGIGIVPAVEPL----IAA 57
R GV+IH ++ + + +++G S D V+V +G VP V L +
Sbjct 241 RKAGVQIHPHSVAKAVRQEADKSLTLELTNGESFRGFDSVIVSVGRVPEVANLGLDVVGV 300
Query 58 GAASGNGVEVDEFCQTSLPNIYAIGDCA--IHANP--FAGGQRI 97
G + DEF TS+ IYA+GD + I P A G+R+
Sbjct 301 KQRHGGYIVADEFQNTSVEQIYAVGDVSGKIELTPVAIAAGRRL 344
> pfa:PF07_0085 ferrodoxin reductase-like protein
Length=642
Score = 35.0 bits (79), Expect = 0.059, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 33/60 (55%), Gaps = 1/60 (1%)
Query 25 VTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVDE-FCQTSLPNIYAIGD 83
+ GVR+++G V+ D V+ +G +P + L N +EVD+ F + N+YA GD
Sbjct 450 IHGVRLNNGEVINCDYVIEALGCIPNSDFLDEKYKNVNNFIEVDKHFKVKNSDNMYAAGD 509
> cel:C26D10.3 hypothetical protein
Length=451
Score = 35.0 bits (79), Expect = 0.063, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 22/25 (88%), Gaps = 1/25 (4%)
Query 63 NGVEVDEFCQTSLPNIYAIGD-CAI 86
+G++V++ C+TSLPN++A GD CA+
Sbjct 312 SGIKVNDACETSLPNVFACGDVCAL 336
> pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=500
Score = 34.7 bits (78), Expect = 0.078, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 28 VRMSDGSVLPA-DMVVVGIGIVPAVEPLIAAG---AASGNGVEVDEFCQTSLPNIYAIGD 83
+ +SDG + D V+ +G P E L + N + VDE +TS+ NIYA+GD
Sbjct 253 IHLSDGRIYEHFDHVIYCVGRSPDTENLNLEKLNVETNNNYIVVDENQRTSVNNIYAVGD 312
Query 84 CAI 86
C +
Sbjct 313 CCM 315
> tgo:TGME49_088830 pyridine nucleotide-disulphide oxidoreductase,
putative (EC:1.6.5.3); K03885 NADH dehydrogenase [EC:1.6.99.3]
Length=657
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 29/54 (53%), Gaps = 7/54 (12%)
Query 44 GIGIVPAVEPLIAA------GAASGNGVEVD-EFCQTSLPNIYAIGDCAIHANP 90
G+G VP V+ +IA G G+ VD + + PN+YA+GDCA A P
Sbjct 424 GVGEVPLVKKIIAENFPNVEGKPRLRGLPVDAQLRLLNQPNVYALGDCAAIAPP 477
> cel:Y56A3A.32 wah-1; Worm AIF (apoptosis inducing factor) Homolog
family member (wah-1); K04727 programmed cell death 8
(apoptosis-inducing factor) [EC:1.-.-.-]
Length=700
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 40/81 (49%), Gaps = 6/81 (7%)
Query 28 VRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGN---GVEVDEFCQTSLPNIYAIGDC 84
+++SDGS L D+VVV G P E + A+G GV D+ C N++A G
Sbjct 490 LKLSDGSELRTDLVVVATGEEPNSEIIEASGLKIDEKLGGVRADK-CLKVGENVWAAGAI 548
Query 85 AIHANPFAGGQRIRLESVQNA 105
A + G +R+ S +NA
Sbjct 549 ATFEDGVLGARRV--SSWENA 567
> eco:b1109 ndh, ECK1095, JW1095; respiratory NADH dehydrogenase
2/cupric reductase (EC:1.16.1.- 1.6.99.3); K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=434
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 35/71 (49%), Gaps = 5/71 (7%)
Query 27 GVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVE---VDEFCQTSL-PNIYAIG 82
G+ DG + AD++V GI A + L G N + V+ QT+ P+IYAIG
Sbjct 256 GLHTKDGEYIEADLMVWAAGI-KAPDFLKDIGGLETNRINQLVVEPTLQTTRDPDIYAIG 314
Query 83 DCAIHANPFAG 93
DCA P G
Sbjct 315 DCASCPRPEGG 325
> eco:b4154 frdA, ECK4150, JW4115; fumarate reductase (anaerobic)
catalytic and NAD/flavoprotein subunit (EC:1.3.99.1); K00244
fumarate reductase flavoprotein subunit [EC:1.3.99.1]
Length=602
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 45 IGIVPAVEPLIAAGAA--SGNGVEVDEFCQTSLPNIYAIGDCA 85
+G+ P EP+ A + G+E D+ C+T + ++A+G+C+
Sbjct 340 VGVDPVKEPIPVRPTAHYTMGGIETDQNCETRIKGLFAVGECS 382
> ath:AT4G21490 NDB3; NDB3; NADH dehydrogenase; K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=580
Score = 32.0 bits (71), Expect = 0.51, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 34 SVLPADMVV--VGIGIVPAVEPLIAA-GAASGNGVEVDEFCQTS-LPNIYAIGDCA 85
S +P M+V GIG P ++ + G + + DE+ + NIYA+GDCA
Sbjct 316 STIPYGMIVWSTGIGTRPVIKDFMKQIGQGNRRALATDEWLRVEGCDNIYALGDCA 371
> ath:AT4G05020 NDB2; NDB2 (NAD(P)H dehydrogenase B2); FAD binding
/ disulfide oxidoreductase/ oxidoreductase; K03885 NADH
dehydrogenase [EC:1.6.99.3]
Length=582
Score = 31.2 bits (69), Expect = 0.79, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 34 SVLPADMVV--VGIGIVPAVEPLIAA-GAASGNGVEVDEFCQTS-LPNIYAIGDCA 85
S +P M+V GIG P ++ + G + + DE+ + NIYA+GDCA
Sbjct 318 SSIPYGMIVWSTGIGTRPVIKDFMKQIGQGNRRALATDEWLRVEGTDNIYALGDCA 373
> dre:373125 pdcd8, AIF, zgc:91994; programmed cell death 8 (apoptosis-inducing
factor); K04727 programmed cell death 8 (apoptosis-inducing
factor) [EC:1.-.-.-]
Length=613
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Query 28 VRMSDGSVLPADMVVVGIGIVPAVEPLIAAGA---ASGNGVEVDEFCQTSLPNIYAIGDC 84
+++ DG ++ D +V +G+ P+VE +AG + G V+ Q NI+ GD
Sbjct 383 IKLKDGRLVKTDHIVAAVGLEPSVELAKSAGLEVDSDFGGYRVNAELQAR-SNIWVAGDA 441
Query 85 AIHANPFAGGQRI 97
A + G +R+
Sbjct 442 ACFYDIKLGRRRV 454
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 25 VTGVRMSDGSVLPADMVVVGIGIVPAVEPL----IAAGAASGNGVEVDEFCQTSLPNIYA 80
V G + L D ++V +G P E L + + V V+E QT +P+I+A
Sbjct 278 VEGAKDGKKQTLECDTLLVSVGRRPYTEGLGLSNVQIDLDNRGRVPVNERFQTKVPSIFA 337
Query 81 IGD 83
IGD
Sbjct 338 IGD 340
> ath:AT2G29990 NDA2; NDA2 (ALTERNATIVE NAD(P)H DEHYDROGENASE
2); FAD binding / NADH dehydrogenase/ oxidoreductase; K03885
NADH dehydrogenase [EC:1.6.99.3]
Length=508
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 34/59 (57%), Gaps = 4/59 (6%)
Query 30 MSDGSVLPADMVV--VGIGIVPAVEPLIAAGAASGNGVEVDEFCQT-SLPNIYAIGDCA 85
+ DG+ +P ++V G+G P V L +G + +DE+ + S+ +++AIGDC+
Sbjct 325 LDDGTEVPYGLLVWSTGVGPSPFVRSLGLPKDPTGR-IGIDEWMRVPSVQDVFAIGDCS 382
> eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide-disulfide
oxidoreductase
Length=441
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 44/87 (50%), Gaps = 6/87 (6%)
Query 3 RAHGVEIHLNAKMDCLIGHDGWVTGVRMSDGSVLPADMVVVGIGIVPA---VEPLIAAGA 59
R GV+I LNA ++ + H+ V S+ + L D +++ G PA + P A A
Sbjct 210 RDQGVDIILNAHVERISHHENQVQV--HSEHAQLAVDALLIASGRQPATASLHPENAGIA 267
Query 60 ASGNG-VEVDEFCQTSLPNIYAIGDCA 85
+ G + VD+ T+ NI+A+GD
Sbjct 268 VNERGAIVVDKRLHTTADNIWAMGDVT 294
> dre:100332074 programmed cell death 8 (apoptosis-inducing factor)-like
Length=750
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Query 28 VRMSDGSVLPADMVVVGIGIVPAVEPLIAAGA---ASGNGVEVDEFCQTSLPNIYAIGDC 84
+++ DG ++ D +V +G+ P+VE +AG + G V+ Q NI+ GD
Sbjct 520 IKLKDGRLVKTDHIVAAVGLEPSVELAKSAGLEVDSDFGGYRVNAELQAR-SNIWVAGDA 578
Query 85 AIHANPFAGGQRI 97
A + G +R+
Sbjct 579 ACFYDIKLGRRRV 591
> xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 28/57 (49%), Gaps = 12/57 (21%)
Query 35 VLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVD--------EFCQTSLPNIYAIGD 83
V+ D+++V IG P E L G+E+D QT +PNIYAIGD
Sbjct 303 VITCDVLLVCIGRRPFTENL----GLQELGIELDNRGRIPINSRFQTKIPNIYAIGD 355
> ath:AT4G28220 NDB1; NDB1 (NAD(P)H dehydrogenase B1); NADH dehydrogenase/
disulfide oxidoreductase; K03885 NADH dehydrogenase
[EC:1.6.99.3]
Length=571
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 22/44 (50%), Gaps = 2/44 (4%)
Query 44 GIGIVPAVEPLI-AAGAASGNGVEVDEFCQ-TSLPNIYAIGDCA 85
G+G P + + G V +E+ Q T N+YA+GDCA
Sbjct 323 GVGTRPVISDFMEQVGQGGRRAVATNEWLQVTGCENVYAVGDCA 366
> tpv:TP03_0021 hypothetical protein
Length=727
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 14/24 (58%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 35 VLPADMVVVGIGIVPAVEPLIAAG 58
V P MV+VG+G+V AV P IA G
Sbjct 494 VSPVIMVLVGMGVVYAVYPAIAPG 517
> eco:b0606 ahpF, ECK0600, JW0599; alkyl hydroperoxide reductase,
F52a subunit, FAD/NAD(P)-binding (EC:1.11.1.15); K03387
alkyl hydroperoxide reductase subunit F [EC:1.6.4.-]
Length=521
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 42/84 (50%), Gaps = 6/84 (7%)
Query 7 VEIHLNAKMDCLIGHDGWVTGVRMSD---GSVLPADM--VVVGIGIVPAVEPLIAAGAAS 61
V+I LNA+ + G V G+ D G + ++ + V IG++P L A +
Sbjct 406 VDIILNAQTTEVKGDGSKVVGLEYRDRVSGDIHNIELAGIFVQIGLLPNTNWLEGAVERN 465
Query 62 GNG-VEVDEFCQTSLPNIYAIGDC 84
G + +D C+T++ ++A GDC
Sbjct 466 RMGEIIIDAKCETNVKGVFAAGDC 489
> cel:C46F11.2 hypothetical protein
Length=473
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 21/82 (25%), Positives = 39/82 (47%), Gaps = 6/82 (7%)
Query 7 VEIHLNAKMDCLI-GHDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPL----IAAGAAS 61
+ +H N ++ +I G DG +T ++ + G + ++ IG P + L +
Sbjct 240 LHLHKNTQVTEVIKGDDGLLT-IKTTTGVIEKVQTLIWAIGRDPLTKELNLERVGVKTDK 298
Query 62 GNGVEVDEFCQTSLPNIYAIGD 83
+ VDE+ TS P I ++GD
Sbjct 299 SGHIIVDEYQNTSAPGILSVGD 320
> bbo:BBOV_III007660 17.m07668; hypothetical protein; K08874 transformation/transcription
domain-associated protein
Length=3963
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 19/42 (45%), Gaps = 0/42 (0%)
Query 62 GNGVEVDEFCQTSLPNIYAIGDCAIHANPFAGGQRIRLESVQ 103
GNG++V+ + IY+ C NP GG + + Q
Sbjct 1582 GNGIKVNRIIANAFLEIYSSDKCGSTLNPILGGSAFNMRNYQ 1623
> cpv:cgd4_580 hypothetical protein
Length=746
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 13/54 (24%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 21 HDGWVTGVRMSDGSVLPADMVVVGIGIVPAVEPLIAAGAASGNGVEVDEFCQTS 74
H +V+G+R+ DG+V+ +MV V + ++ + G + + +++ +F + S
Sbjct 160 HKEFVSGIRLEDGTVVHYNMVKVDEEFLQSLSEHMKVGVSESDFIKMIDFMEKS 213
> hsa:84883 AIFM2, AMID, DKFZp686L1298, PRG3, RP11-367H5.2; apoptosis-inducing
factor, mitochondrion-associated, 2
Length=373
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 30/62 (48%), Gaps = 4/62 (6%)
Query 28 VRMSDGSVLPADMVVVGIGI---VPAVEPLIAAGAASGNGVEVDEFCQTS-LPNIYAIGD 83
V+ G+ + ++V++ GI A + AS + V+E Q N+YAIGD
Sbjct 226 VQTDKGTEVATNLVILCTGIKINSSAYRKAFESRLASSGALRVNEHLQVEGHSNVYAIGD 285
Query 84 CA 85
CA
Sbjct 286 CA 287
> tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=519
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 26/53 (49%), Gaps = 5/53 (9%)
Query 36 LPADMVVVGIGIVP-----AVEPLIAAGAASGNGVEVDEFCQTSLPNIYAIGD 83
+ AD+V+V +G P +E L G V D FC + PNI AIGD
Sbjct 312 MEADVVLVAVGRRPYTKNLGLEELGIETDRVGRVVVDDRFCVPNYPNIRAIGD 364
Lambda K H
0.319 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2057481480
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40