bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2094_orf1
Length=51
Score E
Sequences producing significant alignments: (Bits) Value
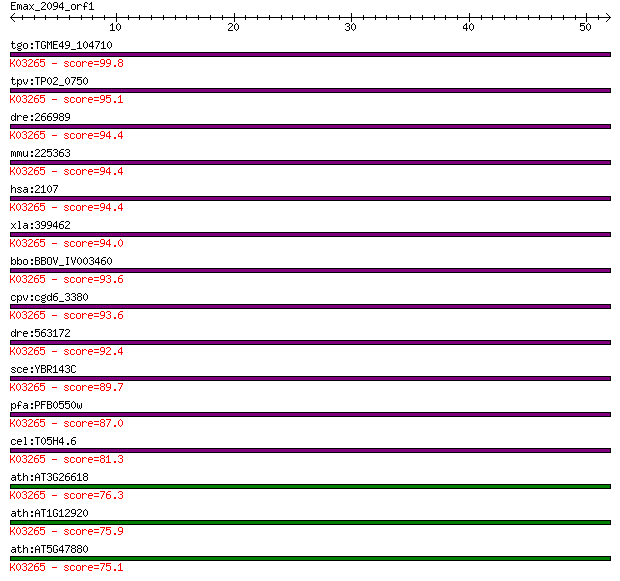
tgo:TGME49_104710 eukaryotic peptide chain release factor subu... 99.8 2e-21
tpv:TP02_0750 eukaryotic peptide chain release factor; K03265 ... 95.1 4e-20
dre:266989 etf1, eRF1, hm:zeh0057, wu:fa19e07, wu:fi31f01, zeh... 94.4 8e-20
mmu:225363 Etf1, AI463371, D6Ertd109e, ERF, ERF1, MGC18745, SU... 94.4 9e-20
hsa:2107 ETF1, D5S1995, ERF, ERF1, MGC111066, RF1, SUP45L1, TB... 94.4 9e-20
xla:399462 etf1; eukaryotic translation termination factor 1; ... 94.0 1e-19
bbo:BBOV_IV003460 21.m02966; peptide chain release factor subu... 93.6 1e-19
cpv:cgd6_3380 Erf1 eukaryotic translation termination factor 1... 93.6 2e-19
dre:563172 zgc:171591; K03265 peptide chain release factor sub... 92.4 3e-19
sce:YBR143C SUP45, SAL4, SUP1, SUP47; Sup45p; K03265 peptide c... 89.7 2e-18
pfa:PFB0550w peptide chain release factor subunit 1, putative;... 87.0 1e-17
cel:T05H4.6 hypothetical protein; K03265 peptide chain release... 81.3 8e-16
ath:AT3G26618 ERF1-3; ERF1-3 (eukaryotic release factor 1-3); ... 76.3 2e-14
ath:AT1G12920 ERF1-2; ERF1-2 (EUKARYOTIC RELEASE FACTOR 1-2); ... 75.9 3e-14
ath:AT5G47880 ERF1-1; ERF1-1 (EUKARYOTIC RELEASE FACTOR 1-1); ... 75.1 5e-14
> tgo:TGME49_104710 eukaryotic peptide chain release factor subunit,
putative ; K03265 peptide chain release factor subunit
1
Length=430
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/51 (90%), Positives = 50/51 (98%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSA+RFARLR+EKRHNY+RKVAETAVQMFITQDKVNVS L+LAGSADF
Sbjct 181 RGGQSAMRFARLRLEKRHNYVRKVAETAVQMFITQDKVNVSGLILAGSADF 231
> tpv:TP02_0750 eukaryotic peptide chain release factor; K03265
peptide chain release factor subunit 1
Length=439
Score = 95.1 bits (235), Expect = 4e-20, Method: Composition-based stats.
Identities = 45/51 (88%), Positives = 48/51 (94%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNYIRKVAETAV MFIT DKVNV+ L+LAG+ADF
Sbjct 187 RGGQSALRFARLRMEKRHNYIRKVAETAVNMFITNDKVNVTGLILAGNADF 237
> dre:266989 etf1, eRF1, hm:zeh0057, wu:fa19e07, wu:fi31f01, zeh0057;
eukaryotic translation termination factor 1; K03265
peptide chain release factor subunit 1
Length=442
Score = 94.4 bits (233), Expect = 8e-20, Method: Composition-based stats.
Identities = 44/51 (86%), Positives = 49/51 (96%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETAVQ+F++ DKVNV+ LVLAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAETAVQLFVSNDKVNVAGLVLAGSADF 232
> mmu:225363 Etf1, AI463371, D6Ertd109e, ERF, ERF1, MGC18745,
SUP45L1, TB3-1; eukaryotic translation termination factor 1;
K03265 peptide chain release factor subunit 1
Length=437
Score = 94.4 bits (233), Expect = 9e-20, Method: Composition-based stats.
Identities = 45/51 (88%), Positives = 49/51 (96%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETAVQ+FI+ DKVNV+ LVLAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAETAVQLFISGDKVNVAGLVLAGSADF 232
> hsa:2107 ETF1, D5S1995, ERF, ERF1, MGC111066, RF1, SUP45L1,
TB3-1; eukaryotic translation termination factor 1; K03265 peptide
chain release factor subunit 1
Length=437
Score = 94.4 bits (233), Expect = 9e-20, Method: Composition-based stats.
Identities = 45/51 (88%), Positives = 49/51 (96%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETAVQ+FI+ DKVNV+ LVLAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAETAVQLFISGDKVNVAGLVLAGSADF 232
> xla:399462 etf1; eukaryotic translation termination factor 1;
K03265 peptide chain release factor subunit 1
Length=437
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 45/51 (88%), Positives = 49/51 (96%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETAVQ+FI+ DKVNV+ LVLAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAETAVQLFISGDKVNVAGLVLAGSADF 232
> bbo:BBOV_IV003460 21.m02966; peptide chain release factor subunit
1; K03265 peptide chain release factor subunit 1
Length=430
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/51 (86%), Positives = 47/51 (92%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRME+RHNY+RKVAE AV MFIT DKVNVS L+LAGSADF
Sbjct 178 RGGQSALRFARLRMERRHNYVRKVAENAVLMFITNDKVNVSGLILAGSADF 228
> cpv:cgd6_3380 Erf1 eukaryotic translation termination factor
1; N-terminal RNAseH plus pelota domain containing protein
; K03265 peptide chain release factor subunit 1
Length=432
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 43/51 (84%), Positives = 47/51 (92%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETA MFIT D+VNV+ L+LAGSADF
Sbjct 181 RGGQSALRFARLRMEKRHNYLRKVAETATTMFITNDQVNVAALILAGSADF 231
> dre:563172 zgc:171591; K03265 peptide chain release factor subunit
1
Length=451
Score = 92.4 bits (228), Expect = 3e-19, Method: Composition-based stats.
Identities = 43/51 (84%), Positives = 48/51 (94%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETAVQ++++ DKVNV LVLAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAETAVQLYVSNDKVNVVGLVLAGSADF 232
> sce:YBR143C SUP45, SAL4, SUP1, SUP47; Sup45p; K03265 peptide
chain release factor subunit 1
Length=437
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 43/51 (84%), Positives = 45/51 (88%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLR EKRHNY+RKVAE AVQ FIT DKVNV L+LAGSADF
Sbjct 179 RGGQSALRFARLREEKRHNYVRKVAEVAVQNFITNDKVNVKGLILAGSADF 229
> pfa:PFB0550w peptide chain release factor subunit 1, putative;
K03265 peptide chain release factor subunit 1
Length=427
Score = 87.0 bits (214), Expect = 1e-17, Method: Composition-based stats.
Identities = 41/51 (80%), Positives = 46/51 (90%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLR+EKRHNY+RKVAE A +FIT DKVNV+ +VLAGSADF
Sbjct 178 RGGQSALRFARLRLEKRHNYVRKVAEVATSVFITNDKVNVTGIVLAGSADF 228
> cel:T05H4.6 hypothetical protein; K03265 peptide chain release
factor subunit 1
Length=443
Score = 81.3 bits (199), Expect = 8e-16, Method: Composition-based stats.
Identities = 38/51 (74%), Positives = 44/51 (86%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSA+RFARLR EKRHNY+RKVAE +V+ FI DKV V+ L+LAGSADF
Sbjct 190 RGGQSAVRFARLRNEKRHNYVRKVAENSVEQFIKNDKVTVAGLILAGSADF 240
> ath:AT3G26618 ERF1-3; ERF1-3 (eukaryotic release factor 1-3);
translation release factor; K03265 peptide chain release factor
subunit 1
Length=435
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/53 (71%), Positives = 42/53 (79%), Gaps = 2/53 (3%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFI--TQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RK AE A Q +I + NVS L+LAGSADF
Sbjct 180 RGGQSALRFARLRMEKRHNYVRKTAELATQFYINPATSQPNVSGLILAGSADF 232
> ath:AT1G12920 ERF1-2; ERF1-2 (EUKARYOTIC RELEASE FACTOR 1-2);
translation release factor; K03265 peptide chain release factor
subunit 1
Length=434
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 38/53 (71%), Positives = 42/53 (79%), Gaps = 2/53 (3%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFI--TQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RK AE A Q +I + NVS L+LAGSADF
Sbjct 179 RGGQSALRFARLRMEKRHNYVRKTAELATQFYINPATSQPNVSGLILAGSADF 231
> ath:AT5G47880 ERF1-1; ERF1-1 (EUKARYOTIC RELEASE FACTOR 1-1);
translation release factor; K03265 peptide chain release factor
subunit 1
Length=436
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 38/53 (71%), Positives = 42/53 (79%), Gaps = 2/53 (3%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFI--TQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RK AE A Q +I + NVS L+LAGSADF
Sbjct 181 RGGQSALRFARLRMEKRHNYVRKTAELATQYYINPATSQPNVSGLILAGSADF 233
Lambda K H
0.326 0.135 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026933688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40