bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2057_orf1
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
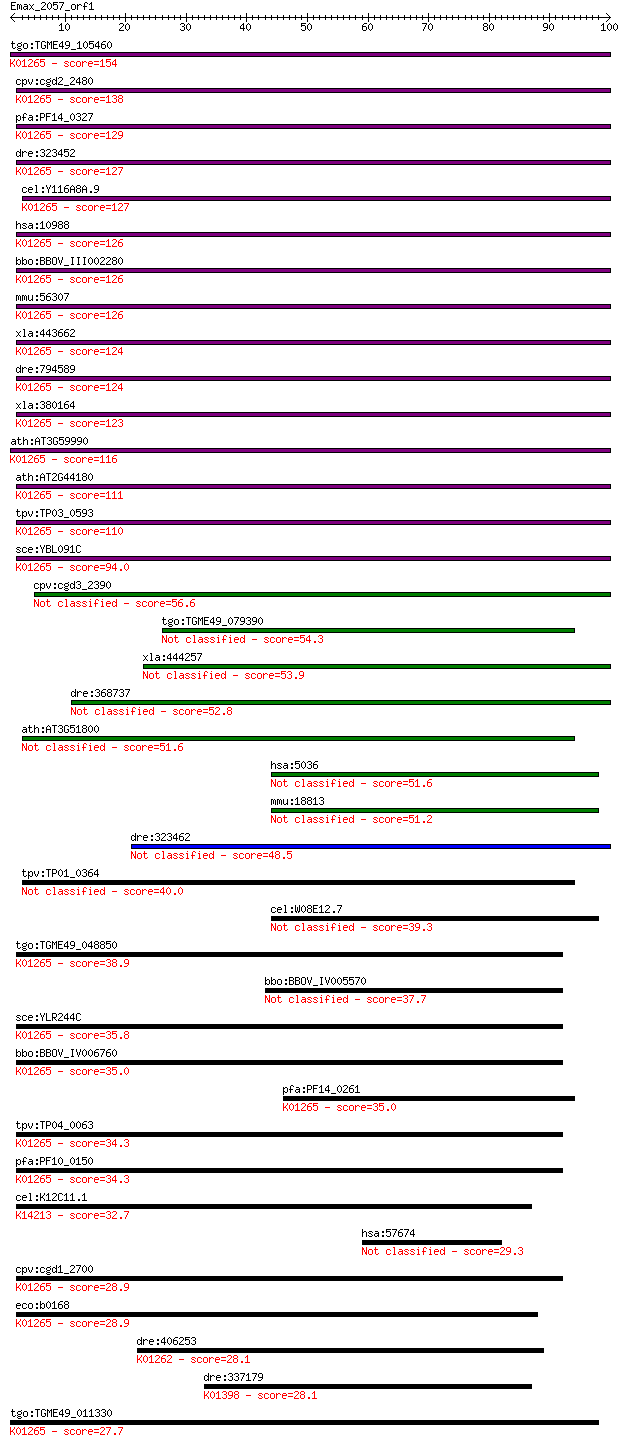
tgo:TGME49_105460 methionine aminopeptidase, type II, putative... 154 7e-38
cpv:cgd2_2480 methionine aminopeptidase, type II, an1 domain ;... 138 4e-33
pfa:PF14_0327 methionine aminopeptidase, type II, putative; K0... 129 3e-30
dre:323452 metap2, wu:fb98h06, zgc:66250; methionyl aminopepti... 127 7e-30
cel:Y116A8A.9 map-2; Methionine AminoPeptidase family member (... 127 9e-30
hsa:10988 METAP2, MAP2, MNPEP, p67, p67eIF2; methionyl aminope... 126 1e-29
bbo:BBOV_III002280 17.m07219; methionine aminopeptidase, type ... 126 1e-29
mmu:56307 Metap2, 4930584B20Rik, A930035J23Rik, AI047573, AL02... 126 1e-29
xla:443662 metap2; methionyl aminopeptidase 2 (EC:3.4.11.18); ... 124 5e-29
dre:794589 metap2l; methionyl aminopeptidase 2 like (EC:3.4.11... 124 1e-28
xla:380164 metap2, MGC53792; methionine aminopeptidase 2 (EC:3... 123 2e-28
ath:AT3G59990 MAP2B; MAP2B (METHIONINE AMINOPEPTIDASE 2B); ami... 116 2e-26
ath:AT2G44180 MAP2A; MAP2A (METHIONINE AMINOPEPTIDASE 2A); ami... 111 5e-25
tpv:TP03_0593 methionine aminopeptidase, type II (EC:3.4.11.18... 110 9e-25
sce:YBL091C MAP2; Map2p (EC:3.4.11.18); K01265 methionyl amino... 94.0 1e-19
cpv:cgd3_2390 proliferation-associated protein 2G4 metalloprot... 56.6 2e-08
tgo:TGME49_079390 proliferation-associated protein 2G4, putati... 54.3 8e-08
xla:444257 pa2g4, ebp1, hg4-1, p38-2g4; proliferation-associat... 53.9 1e-07
dre:368737 pa2g4a, pa2g4, si:dz150i12.2, wu:fb19b11, wu:ft56d0... 52.8 2e-07
ath:AT3G51800 ATG2; ATG2; aminopeptidase/ metalloexopeptidase 51.6 6e-07
hsa:5036 PA2G4, EBP1, HG4-1, p38-2G4; proliferation-associated... 51.6 7e-07
mmu:18813 Pa2g4, 38kDa, AA672939, Ebp1, Plfap; proliferation-a... 51.2 7e-07
dre:323462 pa2g4b, pa2g4l, wu:fb37h04, wu:fb99a12, zgc:65848; ... 48.5 5e-06
tpv:TP01_0364 proliferation-associated protein 2g4 40.0
cel:W08E12.7 hypothetical protein 39.3 0.003
tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.... 38.9 0.004
bbo:BBOV_IV005570 23.m06491; proliferation-associated protein 2g4 37.7
sce:YLR244C MAP1; Map1p (EC:3.4.11.18); K01265 methionyl amino... 35.8 0.035
bbo:BBOV_IV006760 23.m06237; methionine aminopeptidase (EC:3.4... 35.0 0.055
pfa:PF14_0261 proliferation-associated protein 2g4, putative; ... 35.0 0.055
tpv:TP04_0063 methionine aminopeptidase, type I (EC:3.4.11.18)... 34.3 0.099
pfa:PF10_0150 methionine aminopeptidase, putative; K01265 meth... 34.3 0.11
cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase ... 32.7 0.33
hsa:57674 RNF213, C17orf27, DKFZp762N1115, FLJ13051, KIAA1554,... 29.3 3.5
cpv:cgd1_2700 methionine aminopeptidase with MYND finger at N-... 28.9 3.9
eco:b0168 map, ECK0166, JW0163, pepM; methionine aminopeptidas... 28.9 4.5
dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366, ... 28.1 7.8
dre:337179 mmp2, fa99h12, fk89d01, wu:fa99h12, wu:fk89d01; mat... 28.1 8.1
tgo:TGME49_011330 methionine aminopeptidase, putative (EC:3.4.... 27.7 9.4
> tgo:TGME49_105460 methionine aminopeptidase, type II, putative
(EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=480
Score = 154 bits (389), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 67/99 (67%), Positives = 82/99 (82%), Gaps = 0/99 (0%)
Query 1 GLREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINE 60
+R+AAECHRQVRRY Q + RPG+ L +LC LE K++ELI A ++RG GFPTGCS+N
Sbjct 163 AIRQAAECHRQVRRYIQGVARPGVKLVDLCRSLEAKSKELIAAHKLDRGWGFPTGCSLNN 222
Query 61 CAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
CAAHYTPNPG++++L +GDICKLDFGVQV GRIIDCA S
Sbjct 223 CAAHYTPNPGDNRVLEQGDICKLDFGVQVGGRIIDCAFS 261
> cpv:cgd2_2480 methionine aminopeptidase, type II, an1 domain
; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=472
Score = 138 bits (348), Expect = 4e-33, Method: Composition-based stats.
Identities = 60/98 (61%), Positives = 75/98 (76%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR AAE HRQVR+Y QS++RP + L ++C LE K +EL+ A+G++ G GFPTGCS+N C
Sbjct 160 LRRAAEVHRQVRKYMQSIIRPEMKLIDMCNILESKVKELVAAEGLKCGWGFPTGCSLNHC 219
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPNP + L + DICKLDFGVQV G IIDCA +
Sbjct 220 AAHYTPNPHDFTKLTQDDICKLDFGVQVNGMIIDCAFT 257
> pfa:PF14_0327 methionine aminopeptidase, type II, putative;
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=628
Score = 129 bits (323), Expect = 3e-30, Method: Composition-based stats.
Identities = 55/98 (56%), Positives = 75/98 (76%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR+AAECHRQVR++ Q+ ++PG + ++ + E KT+ELI A+ ++ G GFPTGCS+N C
Sbjct 318 LRKAAECHRQVRKHMQAFIKPGKKMIDIAQETERKTKELILAEKLKCGWGFPTGCSLNHC 377
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G++ +L D+CKLDFGV V G IIDCA +
Sbjct 378 AAHYTPNYGDETVLKYDDVCKLDFGVHVNGYIIDCAFT 415
> dre:323452 metap2, wu:fb98h06, zgc:66250; methionyl aminopeptidase
2 (EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=476
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 53/98 (54%), Positives = 71/98 (72%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
R+AAE HRQVR+Y QS ++PG+++ E+C KLE + +LI G+ G FPTGCS+N C
Sbjct 167 FRQAAEAHRQVRKYVQSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNHC 226
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L D+CK+DFG + GRIIDCA +
Sbjct 227 AAHYTPNAGDPTVLQYDDVCKIDFGTHINGRIIDCAFT 264
> cel:Y116A8A.9 map-2; Methionine AminoPeptidase family member
(map-2); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=444
Score = 127 bits (319), Expect = 9e-30, Method: Composition-based stats.
Identities = 51/97 (52%), Positives = 73/97 (75%), Gaps = 0/97 (0%)
Query 3 REAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINECA 62
R +AE HRQVR+Y +S ++PG+++ E+C +LE + LI +G+E G FPTGCS+N CA
Sbjct 134 RRSAEAHRQVRKYVKSWIKPGMTMIEICERLETTSRRLIKEQGLEAGLAFPTGCSLNHCA 193
Query 63 AHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AHYTPN G+ +L GD+CK+D+G+ VRGR+ID A +
Sbjct 194 AHYTPNAGDTTVLQYGDVCKIDYGIHVRGRLIDSAFT 230
> hsa:10988 METAP2, MAP2, MNPEP, p67, p67eIF2; methionyl aminopeptidase
2 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=478
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 54/98 (55%), Positives = 70/98 (71%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
REAAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCS+N C
Sbjct 169 FREAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNNC 228
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L DICK+DFG + GRIIDCA +
Sbjct 229 AAHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFT 266
> bbo:BBOV_III002280 17.m07219; methionine aminopeptidase, type
II family protein; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=432
Score = 126 bits (317), Expect = 1e-29, Method: Composition-based stats.
Identities = 57/98 (58%), Positives = 72/98 (73%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR AAE HRQVRRY QS++RP +SL +L +E K++ELI + G++ G FPTG SIN C
Sbjct 122 LRRAAEVHRQVRRYIQSVIRPDISLIDLVNAIETKSKELIASDGLKCGWAFPTGVSINHC 181
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L GD+CK+DFG V G IIDCA +
Sbjct 182 AAHYTPNYGDKSMLRYGDVCKVDFGTHVNGHIIDCAFT 219
> mmu:56307 Metap2, 4930584B20Rik, A930035J23Rik, AI047573, AL024412,
AU014659, Amp2, MGC102452, Mnpep, p67, p67eIF2; methionine
aminopeptidase 2 (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=478
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 54/98 (55%), Positives = 70/98 (71%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
REAAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCS+N C
Sbjct 169 FREAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNNC 228
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L DICK+DFG + GRIIDCA +
Sbjct 229 AAHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFT 266
> xla:443662 metap2; methionyl aminopeptidase 2 (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=481
Score = 124 bits (312), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 70/98 (71%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
R+AAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCS+N C
Sbjct 172 FRQAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLYAGLAFPTGCSLNNC 231
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L D+CK+DFG + GRIIDCA +
Sbjct 232 AAHYTPNAGDPTVLQYDDVCKIDFGTHINGRIIDCAFT 269
> dre:794589 metap2l; methionyl aminopeptidase 2 like (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=468
Score = 124 bits (310), Expect = 1e-28, Method: Composition-based stats.
Identities = 51/98 (52%), Positives = 70/98 (71%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
R+AAE HRQVRRY +S ++PG+++ ++C +LE + LI G+ G FPTGCS+N C
Sbjct 159 FRQAAEAHRQVRRYVKSWIKPGMTMIDICERLEACSRRLIKEDGLNAGLAFPTGCSLNNC 218
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L D+CK+DFG + GRIIDCA +
Sbjct 219 AAHYTPNAGDPTVLRYDDVCKIDFGTHINGRIIDCAFT 256
> xla:380164 metap2, MGC53792; methionine aminopeptidase 2 (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=462
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 51/98 (52%), Positives = 69/98 (70%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
R+AAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCS+N C
Sbjct 153 FRQAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLYAGLAFPTGCSLNNC 212
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L D+CK+DFG + G IIDCA +
Sbjct 213 AAHYTPNAGDPTVLQYDDVCKIDFGTHINGHIIDCAFT 250
> ath:AT3G59990 MAP2B; MAP2B (METHIONINE AMINOPEPTIDASE 2B); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=439
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 48/99 (48%), Positives = 68/99 (68%), Gaps = 0/99 (0%)
Query 1 GLREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINE 60
+R AAE HRQVR+Y +S+++PG+ +T++C LE +LI G++ G FPTGCS+N
Sbjct 129 SVRRAAEVHRQVRKYVRSIVKPGMLMTDICETLENTVRKLISENGLQAGIAFPTGCSLNW 188
Query 61 CAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAH+TPN G+ +L D+ KLDFG + G IIDCA +
Sbjct 189 VAAHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIIDCAFT 227
> ath:AT2G44180 MAP2A; MAP2A (METHIONINE AMINOPEPTIDASE 2A); aminopeptidase/
metalloexopeptidase; K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=441
Score = 111 bits (278), Expect = 5e-25, Method: Composition-based stats.
Identities = 48/98 (48%), Positives = 67/98 (68%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR+AAE HRQVR+Y +S+L+PG+ + +LC LE +LI G++ G FPTGCS+N
Sbjct 132 LRQAAEVHRQVRKYMRSILKPGMLMIDLCETLENTVRKLISENGLQAGIAFPTGCSLNNV 191
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAH+TPN G+ +L D+ KLDFG + G I+D A +
Sbjct 192 AAHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIVDSAFT 229
> tpv:TP03_0593 methionine aminopeptidase, type II (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=452
Score = 110 bits (276), Expect = 9e-25, Method: Composition-based stats.
Identities = 54/117 (46%), Positives = 71/117 (60%), Gaps = 19/117 (16%)
Query 2 LREAAECHRQVRRYAQSLLRPG-------------------LSLTELCTKLEGKTEELIG 42
+R+AAE HRQ RRY QS+++PG LS ++ LE KT+ LI
Sbjct 123 MRKAAEVHRQARRYIQSVIKPGTIYSIHLYRPTYILQIALGLSCLDIVQALEFKTKYLIE 182
Query 43 AKGIERGKGFPTGCSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
++G++ G GFPTGCS+N CAAHYTPN G+ I + D+ KLDFG V G IID A +
Sbjct 183 SQGLKSGWGFPTGCSLNSCAAHYTPNHGDKTIFHKNDVMKLDFGTHVNGYIIDSAFT 239
> sce:YBL091C MAP2; Map2p (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=421
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 44/105 (41%), Positives = 65/105 (61%), Gaps = 7/105 (6%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGI-------ERGKGFPT 54
+R+ AE HR+VRR + + PG+ L ++ +E T + GA+ + +G GFPT
Sbjct 105 VRKGAEIHRRVRRAIKDRIVPGMKLMDIADMIENTTRKYTGAENLLAMEDPKSQGIGFPT 164
Query 55 GCSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
G S+N CAAH+TPN G+ +L D+ K+D+GVQV G IID A +
Sbjct 165 GLSLNHCAAHFTPNAGDKTVLKYEDVMKVDYGVQVNGNIIDSAFT 209
> cpv:cgd3_2390 proliferation-associated protein 2G4 metalloprotease,
creatinase/aminopeptidase fold
Length=381
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 50/103 (48%), Gaps = 8/103 (7%)
Query 5 AAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELI--------GAKGIERGKGFPTGC 56
AAE +Y +L G ++E+C K + EE G + +++G FPT
Sbjct 31 AAEIVNSTLQYVITLCLDGADISEICRKSDSMIEEKSSSVYNKKEGGRKLDKGIAFPTCI 90
Query 57 SINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
S+NE +++P P E L GD+ K+D G + G I C+ S
Sbjct 91 SVNEICGNFSPLPAESLKLKNGDLIKIDLGAHIDGFISICSHS 133
> tgo:TGME49_079390 proliferation-associated protein 2G4, putative
(EC:3.4.11.18)
Length=462
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 28/71 (39%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Query 26 LTELCTKLEGKTEELIGAKGIERGKGFPTGCSINECAAHYTP---NPGEDKILGEGDICK 82
+ E C+K+ K E K +E+G FPT SINE H++P N D++L EGD+ K
Sbjct 118 IVEACSKVYNKKEN---GKKMEKGIAFPTCISINEICGHFSPVEENAETDRVLAEGDVVK 174
Query 83 LDFGVQVRGRI 93
+D G + G I
Sbjct 175 VDLGCHIDGYI 185
> xla:444257 pa2g4, ebp1, hg4-1, p38-2g4; proliferation-associated
2G4, 38kDa
Length=390
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 44/83 (53%), Gaps = 6/83 (7%)
Query 23 GLSLTELCTKLEGKTEELIGA-----KGIERGKGFPTGCSINECAAHYTP-NPGEDKILG 76
G SL LC K + E G K +++G FPT S+N C H++P +D +L
Sbjct 42 GASLLNLCEKGDAMIMEETGKIFKKEKEMKKGIAFPTSISVNNCVCHFSPLKSDQDYLLK 101
Query 77 EGDICKLDFGVQVRGRIIDCALS 99
+GD+ K+D GV V G I + A S
Sbjct 102 DGDLVKIDLGVHVDGFIANVAHS 124
> dre:368737 pa2g4a, pa2g4, si:dz150i12.2, wu:fb19b11, wu:ft56d05,
zgc:86732; proliferation-associated 2G4, a
Length=392
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 48/95 (50%), Gaps = 6/95 (6%)
Query 11 QVRRYAQSLLRPGLSLTELCTKLEGKTEELIGA-----KGIERGKGFPTGCSINECAAHY 65
Q R +PG+S+ LC K + G K +++G FPT S+N C H+
Sbjct 29 QALRAVIEAAKPGVSVLSLCQKGDAFIMAETGKIFKREKDMKKGIAFPTCVSVNNCVCHF 88
Query 66 TPNPGE-DKILGEGDICKLDFGVQVRGRIIDCALS 99
+P + D +L +GD+ K+D GV V G I + A S
Sbjct 89 SPLKSDPDYMLKDGDLVKIDLGVHVDGFISNVAHS 123
> ath:AT3G51800 ATG2; ATG2; aminopeptidase/ metalloexopeptidase
Length=392
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 3 REAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGA------KGIERGKGFPTGC 56
+ AAE + + + +P + ++C K + +E + K IERG FPT
Sbjct 24 KSAAEIVNKALQVVLAECKPKAKIVDICEKGDSFIKEQTASMYKNSKKKIERGVAFPTCI 83
Query 57 SINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRI 93
S+N H++P ++ +L +GD+ K+D G + G I
Sbjct 84 SVNNTVGHFSPLASDESVLEDGDMVKIDMGCHIDGFI 120
> hsa:5036 PA2G4, EBP1, HG4-1, p38-2G4; proliferation-associated
2G4, 38kDa
Length=394
Score = 51.6 bits (122), Expect = 7e-07, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
K +++G FPT S+N C H++P +D IL EGD+ K+D GV V G I + A
Sbjct 68 KEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVA 122
> mmu:18813 Pa2g4, 38kDa, AA672939, Ebp1, Plfap; proliferation-associated
2G4
Length=394
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
K +++G FPT S+N C H++P +D IL EGD+ K+D GV V G I + A
Sbjct 68 KEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVA 122
> dre:323462 pa2g4b, pa2g4l, wu:fb37h04, wu:fb99a12, zgc:65848;
proliferation-associated 2G4, b
Length=394
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 30/85 (35%), Positives = 47/85 (55%), Gaps = 6/85 (7%)
Query 21 RPGLSLTELCTKLEG-----KTEELIGAKGIERGKGFPTGCSINECAAHYTPNPGE-DKI 74
+PG+S+ LC K + ++ K I++G FPT S+N C H++P + D +
Sbjct 40 KPGVSVLSLCEKGDAFIAAETSKVFKKEKEIKKGIAFPTCVSVNNCVCHFSPIKSDPDYM 99
Query 75 LGEGDICKLDFGVQVRGRIIDCALS 99
L +GD+ K+D GV V G I + A S
Sbjct 100 LKDGDLVKIDLGVHVDGFISNVAHS 124
> tpv:TP01_0364 proliferation-associated protein 2g4
Length=383
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 11/100 (11%)
Query 3 REAAECHRQVRRYAQSLLRPGLSLTELC-----TKLEGKTEELIGAK----GIERGKGFP 53
R A+ + + ++PG+S+ LC T LE +T +L K +++G FP
Sbjct 33 RTASNIANAALKNVLAAVKPGVSVKSLCEVGDATMLE-ETNKLYNKKEHGRKVDKGVAFP 91
Query 54 TGCSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRI 93
T S+NE +++P + + EGD+ K+ G + G +
Sbjct 92 TCVSVNELIDYFSPM-DDSLTVKEGDVVKVTLGCHIDGYV 130
> cel:W08E12.7 hypothetical protein
Length=391
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 26/55 (47%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTPNPGEDK-ILGEGDICKLDFGVQVRGRIIDCA 97
K +G PT SI+ C HYTP E +L G + K+D G + G I A
Sbjct 79 KNFTKGIAMPTCISIDNCICHYTPLKSEAPVVLKNGQVVKVDLGTHIDGLIATAA 133
> tgo:TGME49_048850 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=416
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 43/92 (46%), Gaps = 5/92 (5%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGC--SIN 59
LRE R+ YA SL++PG++ E+ K+ + G + FP C S+N
Sbjct 163 LRETCLLGRRALDYAHSLVKPGVTTEEIDAKVHAFIVDNGGYPSPLNYQQFPKSCCTSVN 222
Query 60 ECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
E H P + + L +GDI +D V +G
Sbjct 223 EVICHGIP---DFRPLQDGDIVNIDITVFFKG 251
> bbo:BBOV_IV005570 23.m06491; proliferation-associated protein
2g4
Length=389
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query 43 AKGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFGVQVRG 91
+ IE+G FPT SINE +++P PG ++ +GD+ K+ G + G
Sbjct 80 GRKIEKGIAFPTCVSINEICDNFSPLEPG--AVIADGDLVKVSLGCHIDG 127
> sce:YLR244C MAP1; Map1p (EC:3.4.11.18); K01265 methionyl aminopeptidase
[EC:3.4.11.18]
Length=387
Score = 35.8 bits (81), Expect = 0.035, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 21/100 (21%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGK--------GFP 53
+R+A R+V A + +RPG++ EL +E++ + I+RG FP
Sbjct 138 IRKACMLGREVLDIAAAHVRPGITTDEL--------DEIVHNETIKRGAYPSPLNYYNFP 189
Query 54 TG--CSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
S+NE H P + +L EGDI LD + +G
Sbjct 190 KSLCTSVNEVICHGVP---DKTVLKEGDIVNLDVSLYYQG 226
> bbo:BBOV_IV006760 23.m06237; methionine aminopeptidase (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=376
Score = 35.0 bits (79), Expect = 0.055, Method: Composition-based stats.
Identities = 26/92 (28%), Positives = 43/92 (46%), Gaps = 5/92 (5%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTG--CSIN 59
+R+A+ R+ +A SL+ PG++ E+ TK+ + GFP S+N
Sbjct 130 IRKASILGRKALDFAASLIAPGVTTDEIDTKVHDFIIQHNAYPSPLNYYGFPKSLCTSVN 189
Query 60 ECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
E H P + + L +GDI +D V + G
Sbjct 190 EVVCHGIP---DKRPLKDGDIINIDISVYLNG 218
> pfa:PF14_0261 proliferation-associated protein 2g4, putative;
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=377
Score = 35.0 bits (79), Expect = 0.055, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 46 IERGKGFPTGCSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRI 93
+E+G FP ++NE +Y P+ + + GDI K+ G + G I
Sbjct 69 VEKGISFPVTINVNEICNNYAPSLDCVETIKNGDIVKISLGCHIDGHI 116
> tpv:TP04_0063 methionine aminopeptidase, type I (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=378
Score = 34.3 bits (77), Expect = 0.099, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 40/92 (43%), Gaps = 5/92 (5%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTG--CSIN 59
+R A R+ A SL++PG++ E+ TK+ G FP S+N
Sbjct 129 IRRACLLGRKALDLANSLIKPGITTDEIDTKVHEFIVSHNGYPSPLNYYNFPKSICTSVN 188
Query 60 ECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
E H P+ + L EGDI +D V + G
Sbjct 189 EVVCHGIPDL---RPLEEGDIVNVDISVYLNG 217
> pfa:PF10_0150 methionine aminopeptidase, putative; K01265 methionyl
aminopeptidase [EC:3.4.11.18]
Length=517
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 5/92 (5%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGC--SIN 59
+REA R+ YA +L+ PG++ E+ K+ + FP C S+N
Sbjct 261 IREACILGRKTLDYAHTLVSPGVTTDEIDRKVHEFIIKNNAYPSTLNYYKFPKSCCTSVN 320
Query 60 ECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
E H P + + L GDI +D V +G
Sbjct 321 EIVCHGIP---DYRPLKSGDIINIDISVFYKG 349
> cel:K12C11.1 hypothetical protein; K14213 Xaa-Pro dipeptidase
[EC:3.4.13.9]
Length=498
Score = 32.7 bits (73), Expect = 0.33, Method: Composition-based stats.
Identities = 24/86 (27%), Positives = 37/86 (43%), Gaps = 3/86 (3%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
+R A++ + R A +RPGL +L + + G + + TGC N
Sbjct 192 MRYASKIASEAHRAAMKHMRPGLYEYQLESLFRHTSYYHGGCRHLAYTCIAATGC--NGS 249
Query 62 AAHYT-PNPGEDKILGEGDICKLDFG 86
HY N DK + +GD+C D G
Sbjct 250 VLHYGHANAPNDKFIKDGDMCLFDMG 275
> hsa:57674 RNF213, C17orf27, DKFZp762N1115, FLJ13051, KIAA1554,
KIAA1618, MGC46622, MGC9929, NET57; ring finger protein 213
Length=5256
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 59 NECAAHYTPNPGEDKILGEGDIC 81
N C HYT + G D++L EG +C
Sbjct 465 NICELHYTRDLGHDRVLVEGIVC 487
> cpv:cgd1_2700 methionine aminopeptidase with MYND finger at
N-terminus ; K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=407
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 41/96 (42%), Gaps = 13/96 (13%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGK----GFPTGC- 56
LRE + R+ A S+++PG++ ++ I +K FP C
Sbjct 158 LRECCKIGREALDIAASMIKPGVT----TDAIDEAVHNFIISKNSYPSPLNYWEFPKSCC 213
Query 57 -SINECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
S+NE H P + + L EGDI +D V +G
Sbjct 214 TSVNEIICHGIP---DFRPLEEGDIVNVDISVYYKG 246
> eco:b0168 map, ECK0166, JW0163, pepM; methionine aminopeptidase
(EC:3.4.11.18); K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=264
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 37/89 (41%), Gaps = 5/89 (5%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTEL---CTKLEGKTEELIGAKGIERGKGFPTGCSI 58
+R A +V + ++PG+S EL C + + A G SI
Sbjct 14 MRVAGRLAAEVLEMIEPYVKPGVSTGELDRICNDYIVNEQHAVSACLGYHGYPKSVCISI 73
Query 59 NECAAHYTPNPGEDKILGEGDICKLDFGV 87
NE H P+ + K+L +GDI +D V
Sbjct 74 NEVVCHGIPD--DAKLLKDGDIVNIDVTV 100
> dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366,
zgc:77772; X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
(EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=620
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 4/69 (5%)
Query 22 PGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSI--NECAAHYTPNPGEDKILGEGD 79
P ++TE+ K EEL + G FPT S+ N HY P P ++ L +
Sbjct 353 PKGTVTEISAA--DKAEELRSQQKEFVGLSFPTISSVGPNGAIIHYRPLPETNRTLSLNE 410
Query 80 ICKLDFGVQ 88
+ +D G Q
Sbjct 411 VYLIDSGAQ 419
> dre:337179 mmp2, fa99h12, fk89d01, wu:fa99h12, wu:fk89d01; matrix
metalloproteinase 2 (EC:3.4.24.24); K01398 matrix metalloproteinase-2
(gelatinase A) [EC:3.4.24.24]
Length=657
Score = 28.1 bits (61), Expect = 8.1, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 27/64 (42%), Gaps = 10/64 (15%)
Query 33 LEGKTEELIGAKGIERGKGFPTGCSINECAAHYTPNPG----------EDKILGEGDICK 82
++G+ + +I E G G+P A + P PG E LGEG + K
Sbjct 163 MDGEADIMINFGRNEHGDGYPFDGKDGLLAHAFAPGPGIGGDSHFDDDEQWTLGEGQVVK 222
Query 83 LDFG 86
+ +G
Sbjct 223 VKYG 226
> tgo:TGME49_011330 methionine aminopeptidase, putative (EC:3.4.11.18);
K01265 methionyl aminopeptidase [EC:3.4.11.18]
Length=329
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 43/107 (40%), Gaps = 14/107 (13%)
Query 1 GLREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGK--------GF 52
G+R A E R+V + A ++ G+ + ++ + ++RG F
Sbjct 69 GVRRACEVTREVLQVAVDFVK-GVCAQSSAPLTTEDIDRVVHEETMKRGAYPSPLRYCNF 127
Query 53 PTGC--SINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
P S NE H P+ D+ L G IC +D + G DCA
Sbjct 128 PKSVCTSTNEIVCHGIPD---DRPLQRGSICSIDVSCFLDGFHGDCA 171
Lambda K H
0.319 0.141 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2046143372
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40