bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2034_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
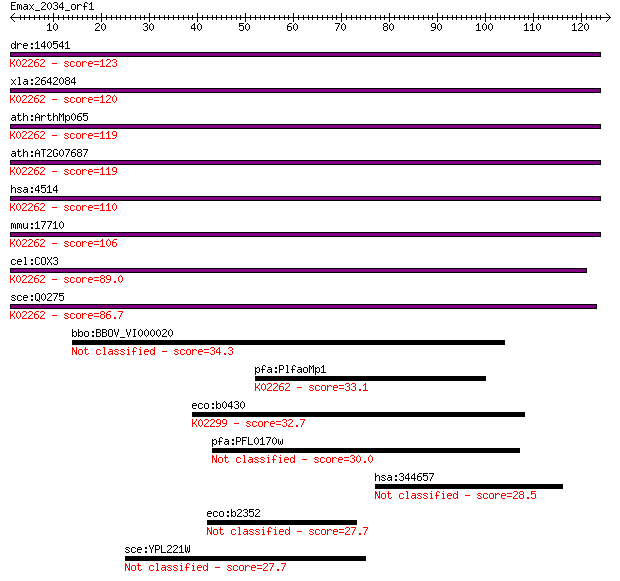
dre:140541 COX3, mtco3; cytochrome c oxidase subunit III; K022... 123 1e-28
xla:2642084 COX3; cytochrome c oxidase subunit III; K02262 cyt... 120 1e-27
ath:ArthMp065 cox3; cytochrome c oxidase subunit 3; K02262 cyt... 119 3e-27
ath:AT2G07687 cytochrome c oxidase subunit 3 (EC:1.9.3.1); K02... 119 3e-27
hsa:4514 COX3, COIII, MTCO3; cytochrome c oxidase III; K02262 ... 110 1e-24
mmu:17710 COX3; cytochrome c oxidase subunit III (EC:1.9.3.1);... 106 2e-23
cel:COX3 cytochrome c oxidase subunit III; K02262 cytochrome c... 89.0 3e-18
sce:Q0275 COX3, OXI2; Cox3p (EC:1.9.3.1); K02262 cytochrome c ... 86.7 2e-17
bbo:BBOV_VI000020 cytochrome c oxidase subunit III 34.3 0.094
pfa:PlfaoMp1 coxIII; putative cytochrome oxidase III; K02262 c... 33.1 0.25
eco:b0430 cyoC, ECK0424, JW0420; cytochrome o ubiquinol oxidas... 32.7 0.29
pfa:PFL0170w transporter, putative 30.0 1.9
hsa:344657 LRRIQ4, LRRC64; leucine-rich repeats and IQ motif c... 28.5 5.3
eco:b2352 gtrS, ECK2346, gtrIV, JW5382, yfdI; serotype-specifi... 27.7 8.5
sce:YPL221W FLC1; Flc1p 27.7 10.0
> dre:140541 COX3, mtco3; cytochrome c oxidase subunit III; K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=261
Score = 123 bits (309), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 58/123 (47%), Positives = 80/123 (65%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP G+ + DP +P LNT L++SGV+VTW HH++ +G+ + I SL LTILLG FT+
Sbjct 117 PPTGLTTLDPFEVPLLNTAVLLASGVTVTWAHHSLMEGERKQAIQSLALTILLGLYFTAL 176
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EYY A FT + Y S +F+ TGFHGLHV+IG L +CL+R + +H GFE
Sbjct 177 QAMEYYEAPFTIADGVYGSTFFVATGFHGLHVIIGSTFLAVCLLRQVLFHFTSDHHFGFE 236
Query 121 CSC 123
+
Sbjct 237 AAA 239
> xla:2642084 COX3; cytochrome c oxidase subunit III; K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=260
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/123 (48%), Positives = 78/123 (63%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI +P +P LNT L++SGV+VTW HH+I GD + I SL LTILLG FT+
Sbjct 117 PPTGITPLNPFEVPLLNTAVLLASGVTVTWAHHSIMHGDRKEAIQSLTLTILLGLYFTAL 176
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EYY A FT + Y S +F+ TGFHGLHV+IG + L +CL+R + H GFE
Sbjct 177 QAMEYYEAPFTIADGVYGSTFFVATGFHGLHVIIGSLFLSVCLLRQIQYHFTSKHHFGFE 236
Query 121 CSC 123
+
Sbjct 237 AAW 239
> ath:ArthMp065 cox3; cytochrome c oxidase subunit 3; K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=265
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 62/123 (50%), Positives = 75/123 (60%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI DP IPFLNT L SSG +VTW HHAI G + + +L T+LL FT F
Sbjct 120 PPKGIEVLDPWEIPFLNTPILPSSGAAVTWAHHAILAGKEKRAVYALVATVLLALVFTGF 179
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EYY A FT S S Y S +F+ TGFHG HV+IG + L IC IR ++ H VGFE
Sbjct 180 QGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHLTKEHHVGFE 239
Query 121 CSC 123
+
Sbjct 240 AAA 242
> ath:AT2G07687 cytochrome c oxidase subunit 3 (EC:1.9.3.1); K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=265
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 62/123 (50%), Positives = 75/123 (60%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI DP IPFLNT L SSG +VTW HHAI G + + +L T+LL FT F
Sbjct 120 PPKGIEVLDPWEIPFLNTPILPSSGAAVTWAHHAILAGKEKRAVYALVATVLLALVFTGF 179
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EYY A FT S S Y S +F+ TGFHG HV+IG + L IC IR ++ H VGFE
Sbjct 180 QGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHLTKEHHVGFE 239
Query 121 CSC 123
+
Sbjct 240 AAA 242
> hsa:4514 COX3, COIII, MTCO3; cytochrome c oxidase III; K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=261
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 55/123 (44%), Positives = 73/123 (59%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI +P +P LNT L++SGVS+TW HH++ + + I +L +TILLG FT
Sbjct 117 PPTGITPLNPLEVPLLNTSVLLASGVSITWAHHSLMENNRNQMIQALLITILLGLYFTLL 176
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q EY+ + FT S Y S +F+ TGFHGLHV+IG L IC IR + H GFE
Sbjct 177 QASEYFESPFTISDGIYGSTFFVATGFHGLHVIIGSTFLTICFIRQLMFHFTSKHHFGFE 236
Query 121 CSC 123
+
Sbjct 237 AAA 239
> mmu:17710 COX3; cytochrome c oxidase subunit III (EC:1.9.3.1);
K02262 cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=261
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 52/123 (42%), Positives = 73/123 (59%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI+ +P +P LNT L++SGVS+TW HH++ +G +L +TI+LG FT
Sbjct 117 PPTGISPLNPLEVPLLNTSVLLASGVSITWAHHSLMEGKRNHMNQALLITIMLGLYFTIL 176
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q EY+ F+ S Y S +F+ TGFHGLHV+IG L +CL+R + H GFE
Sbjct 177 QASEYFETSFSISDGIYGSTFFMATGFHGLHVIIGSTFLIVCLLRQLKFHFTSKHHFGFE 236
Query 121 CSC 123
+
Sbjct 237 AAA 239
> cel:COX3 cytochrome c oxidase subunit III; K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=255
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 72/120 (60%), Gaps = 2/120 (1%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
P G+ +P G+P LNT+ L+SSGV+VTW HH++ ++ S+ LT LL A FT
Sbjct 113 SPFGMHLVNPFGVPLLNTIILLSSGVTVTWAHHSLLSN--KSCTNSMILTCLLAAYFTGI 170
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
QL+EY A F+ + + S++++ TGFHG+HVL G + L +R + NH +G E
Sbjct 171 QLMEYMEASFSIADGVFGSIFYLSTGFHGIHVLCGGLFLAFNFLRLLKNHFNYNHHLGLE 230
> sce:Q0275 COX3, OXI2; Cox3p (EC:1.9.3.1); K02262 cytochrome
c oxidase subunit III [EC:1.9.3.1]
Length=269
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 45/122 (36%), Positives = 70/122 (57%), Gaps = 0/122 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP+GI + P +P LNT+ L+SSG +VT++HHA+ G+ + L +T L F +
Sbjct 125 PPVGIEAVQPTELPLLNTIILLSSGATVTYSHHALIAGNRNKALSGLLITFWLIVIFVTC 184
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EY A FT S Y SV++ GTG H LH+++ +L + R ++ H VG+E
Sbjct 185 QYIEYTNAAFTISDGVYGSVFYAGTGLHFLHMVMLAAMLGVNYWRMRNYHLTAGHHVGYE 244
Query 121 CS 122
+
Sbjct 245 TT 246
> bbo:BBOV_VI000020 cytochrome c oxidase subunit III
Length=228
Score = 34.3 bits (77), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 47/94 (50%), Gaps = 9/94 (9%)
Query 14 PFLN---TVPLVSSGVSVTWTHHAIEQ-GDYRARIISLFLTILLGATFTSFQLLEYYVAG 69
P+LN ++ + SS +S+ + D+ + S F +G+ F S+Q EY
Sbjct 101 PYLNITSSLNITSSVISILVYKMTVSHFSDFEKWLTSAFF---IGSLFLSYQGDEYTFLQ 157
Query 70 FTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICL 103
+ S +S + I TG H LHV +G +LFICL
Sbjct 158 CGLNDSWFSLAFLIITGLHSLHVCVG--VLFICL 189
> pfa:PlfaoMp1 coxIII; putative cytochrome oxidase III; K02262
cytochrome c oxidase subunit III [EC:1.9.3.1]
Length=279
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 52 LLGATFTSFQLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLL 99
LLG F S Q EY + + + Y+++++ TG H HV+IG +LL
Sbjct 175 LLGECFASLQTTEYLHLSYHINDTVYTTLFYCVTGLHFSHVVIGLLLL 222
> eco:b0430 cyoC, ECK0424, JW0420; cytochrome o ubiquinol oxidase
subunit III (EC:1.10.3.-); K02299 cytochrome o ubiquinol
oxidase subunit III [EC:1.10.3.-]
Length=204
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 40/73 (54%), Gaps = 4/73 (5%)
Query 39 DYRARIIS-LFLTILLGATFTSFQLLEYY---VAGFTFSCSAYSSVYFIGTGFHGLHVLI 94
+ ++++IS L LT L GA F ++ E++ V G S + S +F G HGLHV
Sbjct 93 NNKSQVISWLALTWLFGAGFIGMEIYEFHHLIVNGMGPDRSGFLSAFFALVGTHGLHVTS 152
Query 95 GRVLLFICLIRFS 107
G + + + +++ +
Sbjct 153 GLIWMAVLMVQIA 165
> pfa:PFL0170w transporter, putative
Length=1250
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 33/64 (51%), Gaps = 2/64 (3%)
Query 43 RIISLFLTILLGATFTSFQLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFIC 102
+I++LF +L+ TF SF L+ + + F S Y + Y I T L + I V LF+
Sbjct 1084 KILNLFGYLLI--TFVSFSLILFLYSTSYFHISLYDNTYIIYTIIFYLFLFIPNVSLFLF 1141
Query 103 LIRF 106
I +
Sbjct 1142 SINY 1145
> hsa:344657 LRRIQ4, LRRC64; leucine-rich repeats and IQ motif
containing 4
Length=560
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 77 YSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNH 115
++S++ + G GLH L G F CL+ FL +S NH
Sbjct 279 WTSLHLLYLGNTGLHRLRGS---FRCLVNLRFLDLSQNH 314
> eco:b2352 gtrS, ECK2346, gtrIV, JW5382, yfdI; serotype-specific
glucosyl transferase, CPS-53 (KpLE1) prophage
Length=443
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 22/35 (62%), Gaps = 4/35 (11%)
Query 42 ARIISLFLTIL----LGATFTSFQLLEYYVAGFTF 72
AR++SLFL ++ L A FTS+ E YV FTF
Sbjct 92 ARVLSLFLKVIYIYSLYAIFTSYIKTERYVTLFTF 126
> sce:YPL221W FLC1; Flc1p
Length=793
Score = 27.7 bits (60), Expect = 10.0, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 25 GVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSFQLLEYYVAGFTFSC 74
G S T I++ YR +I + T ++ FT F L Y++AGF +C
Sbjct 294 GNSNTLIFRGIKRMGYRMKIEN---TAIVCTGFTFFVLCGYFLAGFIMAC 340
Lambda K H
0.331 0.146 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40