bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2024_orf2
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
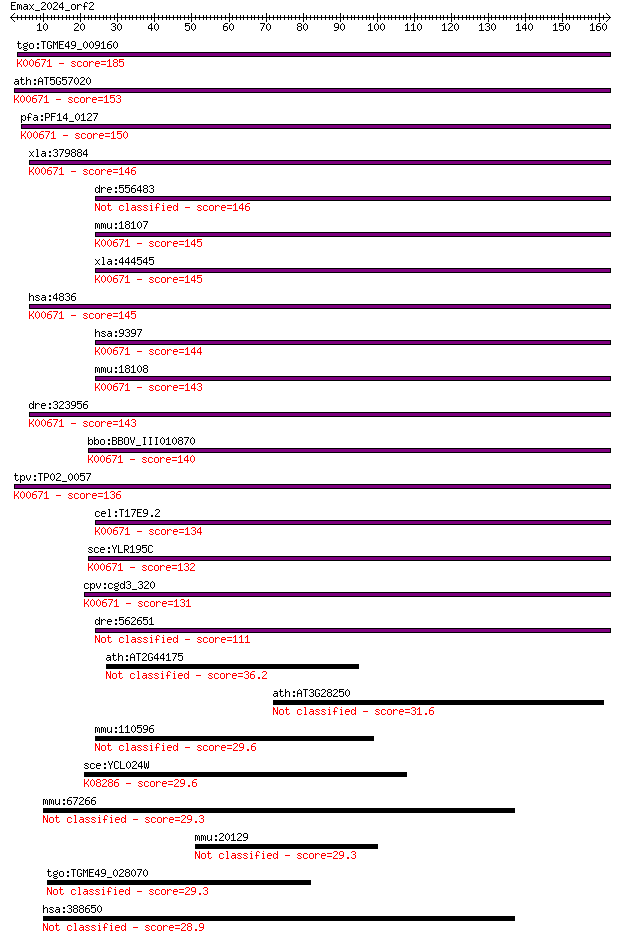
tgo:TGME49_009160 glycylpeptide N-tetradecanoyltransferase, pu... 185 5e-47
ath:AT5G57020 NMT1; NMT1 (MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRA... 153 3e-37
pfa:PF14_0127 N-myristoyltransferase (EC:2.3.1.97); K00671 gly... 150 2e-36
xla:379884 nmt2, MGC53594, nmt1; N-myristoyltransferase 2 (EC:... 146 3e-35
dre:556483 nmt2, fb40e03, wu:fb40e03; N-myristoyltransferase 2... 146 3e-35
mmu:18107 Nmt1, AW536594; N-myristoyltransferase 1 (EC:2.3.1.9... 145 6e-35
xla:444545 nmt1, MGC83363; N-myristoyltransferase 1 (EC:2.3.1.... 145 6e-35
hsa:4836 NMT1, NMT; N-myristoyltransferase 1 (EC:2.3.1.97); K0... 145 6e-35
hsa:9397 NMT2; N-myristoyltransferase 2 (EC:2.3.1.97); K00671 ... 144 1e-34
mmu:18108 Nmt2, A930001K02Rik, AI605445, AU044698, hNMT-2; N-m... 143 2e-34
dre:323956 nmt1a, im:2601337, nmt1, wu:fc15d01, wu:fc18a04, zg... 143 3e-34
bbo:BBOV_III010870 17.m07935; myristoyl-CoA:protein N-myristoy... 140 2e-33
tpv:TP02_0057 N-myristoyltransferase; K00671 glycylpeptide N-t... 136 3e-32
cel:T17E9.2 nmt-1; N-Myristoyl Transferase homolog family memb... 134 2e-31
sce:YLR195C NMT1, CDC72; N-myristoyl transferase, catalyzes th... 132 6e-31
cpv:cgd3_320 N-myristoyltransferase ; K00671 glycylpeptide N-t... 131 1e-30
dre:562651 nmt1b; si:dkeyp-94b4.2 (EC:2.3.1.97) 111 8e-25
ath:AT2G44175 N-myristoyltransferase-related 36.2 0.046
ath:AT3G28250 glycosyl hydrolase family protein 17 31.6
mmu:110596 Rgnef, 9230110L08Rik, AI323540, D13Bwg1089e, RIP2, ... 29.6 4.3
sce:YCL024W KCC4; Kcc4p (EC:2.7.11.1); K08286 protein-serine/t... 29.6 4.6
mmu:67266 Fam69a, 2900024C23Rik, AI315274, MGC144843; family w... 29.3 5.4
mmu:20129 Rptn; repetin 29.3 5.8
tgo:TGME49_028070 hypothetical protein 29.3 6.1
hsa:388650 FAM69A, FLJ23493; family with sequence similarity 6... 28.9 7.0
> tgo:TGME49_009160 glycylpeptide N-tetradecanoyltransferase,
putative (EC:2.3.1.97); K00671 glycylpeptide N-tetradecanoyltransferase
[EC:2.3.1.97]
Length=448
Score = 185 bits (470), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 90/160 (56%), Positives = 114/160 (71%), Gaps = 4/160 (2%)
Query 3 SLVQAIRGLSLSRSPLNSPHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYN 62
SL++ +R + SP +PH FWDTQPV K +E K EGPI+ TKTV+ V+ EPY
Sbjct 63 SLMREMRVSNKGVSPPFAPHTFWDTQPVPKLNEPKE---GKEGPIE-TKTVDQVRSEPYK 118
Query 63 LPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWI 122
LP+GFVW EC V DP+ L E+Y+LL+ HYVEDDDNLFRFNY DFL WAL P ++W+
Sbjct 119 LPDGFVWCECDVRDPEELKEVYDLLSQHYVEDDDNLFRFNYSADFLDWALTAPGCHRDWV 178
Query 123 IGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
IGVRV ++ KLVGFI+A P+ + S + +AEVNFLCVH
Sbjct 179 IGVRVSSTNKLVGFITATPSQIRVFSDSVPMAEVNFLCVH 218
> ath:AT5G57020 NMT1; NMT1 (MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE);
glycylpeptide N-tetradecanoyltransferase/ myristoyltransferase;
K00671 glycylpeptide N-tetradecanoyltransferase
[EC:2.3.1.97]
Length=434
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 72/161 (44%), Positives = 104/161 (64%), Gaps = 2/161 (1%)
Query 2 ASLVQAIRGLSLSRSPLNSPHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPY 61
SL +R S S + H FW+TQPV + + T+ EGPI+ + +VK+EPY
Sbjct 29 TSLETIVRRFQDSMSEAKT-HKFWETQPVGQFKDIGD-TSLPEGPIEPATPLSEVKQEPY 86
Query 62 NLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEW 121
NLP+ + W+ C + + E+YNLL +YVEDD+N+FRFNY K+FL WAL PP +++ W
Sbjct 87 NLPSVYEWTTCDMNSDDMCSEVYNLLKNNYVEDDENMFRFNYSKEFLRWALRPPGYYQSW 146
Query 122 IIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
IGVR + S+KLV FIS +P + + +++AE+NFLCVH
Sbjct 147 HIGVRAKTSKKLVAFISGVPARIRVRDEVVKMAEINFLCVH 187
> pfa:PF14_0127 N-myristoyltransferase (EC:2.3.1.97); K00671 glycylpeptide
N-tetradecanoyltransferase [EC:2.3.1.97]
Length=410
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 74/159 (46%), Positives = 101/159 (63%), Gaps = 5/159 (3%)
Query 4 LVQAIRGLSLSRSPLNSPHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNL 63
L Q IR ++ + + FW TQPV K +++ + P + VEDV+KE Y L
Sbjct 13 LYQLIRN---AKDKIKIDYKFWYTQPVPKINDEFDENVNE--PFISDNKVEDVRKEEYKL 67
Query 64 PNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWII 123
P+G+ W C + ++YNLL +YVEDDDN+FRFNY +FL+WAL+ PN+ K W I
Sbjct 68 PSGYAWCVCDITKENDRSDIYNLLTDNYVEDDDNVFRFNYSSEFLLWALSSPNYVKNWHI 127
Query 124 GVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
GV+ E++ KLVGFISAIP + N I++AEVNFLCVH
Sbjct 128 GVKYESTNKLVGFISAIPIDMCVNKNIIKMAEVNFLCVH 166
> xla:379884 nmt2, MGC53594, nmt1; N-myristoyltransferase 2 (EC:2.3.1.97);
K00671 glycylpeptide N-tetradecanoyltransferase
[EC:2.3.1.97]
Length=484
Score = 146 bits (369), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 73/164 (44%), Positives = 101/164 (61%), Gaps = 13/164 (7%)
Query 6 QAIRGLSLSRSPLNS-------PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKK 58
+AI S+ + P + + FWDTQPV K E GPI+ K +++++
Sbjct 82 KAIELFSVGQGPAKTMEEASKRSYQFWDTQPVPKLGE----VVSSHGPIEPDK--DNIRQ 135
Query 59 EPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
EPY LP GF+W + D VL ELY LL +YVEDDDN+FRF+Y +FL+WAL PP +
Sbjct 136 EPYTLPQGFIWDALDLGDRVVLKELYTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWL 195
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
+W GVRV +S+KLVGFISA+P ++ ++ E+NFLCVH
Sbjct 196 PQWHCGVRVMSSKKLVGFISAVPASMHLYDITKKMVEINFLCVH 239
> dre:556483 nmt2, fb40e03, wu:fb40e03; N-myristoyltransferase
2 (EC:2.3.1.97)
Length=493
Score = 146 bits (368), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 72/139 (51%), Positives = 95/139 (68%), Gaps = 6/139 (4%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K +E +TT GPI+ K E++++EPY+LP GF+W + + +VL EL
Sbjct 116 FWDTQPVPKLNE--VVTTH--GPIEPDK--ENIRQEPYSLPQGFMWDTLDLSNAEVLKEL 169
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
Y LL +YVEDDDN+FRF+Y +FL WAL PP + W GVRV +++KLVGFISAIP
Sbjct 170 YTLLNENYVEDDDNMFRFDYSPNFLKWALRPPGWLPHWHCGVRVSSNKKLVGFISAIPAD 229
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ R+ E+NFLCVH
Sbjct 230 IHIYDTLKRMVEINFLCVH 248
> mmu:18107 Nmt1, AW536594; N-myristoyltransferase 1 (EC:2.3.1.97);
K00671 glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=496
Score = 145 bits (366), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 69/139 (49%), Positives = 90/139 (64%), Gaps = 6/139 (4%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K E GP++ K +++++EPY LP GF W + D VL EL
Sbjct 119 FWDTQPVPKLGE----VVNTHGPVEPDK--DNIRQEPYTLPQGFTWDALDLGDRGVLKEL 172
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
Y LL +YVEDDDN+FRF+Y +FL+WAL PP + +W GVRV +S+KLVGFISAIP
Sbjct 173 YTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWLPQWHCGVRVVSSRKLVGFISAIPAN 232
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ ++ E+NFLCVH
Sbjct 233 IHIYDTEKKMVEINFLCVH 251
> xla:444545 nmt1, MGC83363; N-myristoyltransferase 1 (EC:2.3.1.97);
K00671 glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=498
Score = 145 bits (366), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 68/139 (48%), Positives = 96/139 (69%), Gaps = 6/139 (4%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K +E + GPI+A K +++++EPY+LP GF+W + + +VL EL
Sbjct 121 FWDTQPVPKLNEIITC----HGPIEADK--DNIRQEPYSLPQGFMWDTLDLSNAEVLKEL 174
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
Y LL +YVEDDDN+FRF+Y +FL WAL PP + +W GVRV +++KLVGFISA+P +
Sbjct 175 YTLLNENYVEDDDNMFRFDYSPEFLQWALRPPGWLSQWHCGVRVASNKKLVGFISAVPAS 234
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ ++ E+NFLCVH
Sbjct 235 IRIYDTVKKMVEINFLCVH 253
> hsa:4836 NMT1, NMT; N-myristoyltransferase 1 (EC:2.3.1.97);
K00671 glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=496
Score = 145 bits (366), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 73/164 (44%), Positives = 99/164 (60%), Gaps = 13/164 (7%)
Query 6 QAIRGLSLSRSPLNS-------PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKK 58
+AI S+ + P + + FWDTQPV K E GP++ K +++++
Sbjct 94 KAIELFSVGQGPAKTMEEASKRSYQFWDTQPVPKLGE----VVNTHGPVEPDK--DNIRQ 147
Query 59 EPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
EPY LP GF W + D VL ELY LL +YVEDDDN+FRF+Y +FL+WAL PP +
Sbjct 148 EPYTLPQGFTWDALDLGDRGVLKELYTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWL 207
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
+W GVRV +S+KLVGFISAIP + ++ E+NFLCVH
Sbjct 208 PQWHCGVRVVSSRKLVGFISAIPANIHIYDTEKKMVEINFLCVH 251
> hsa:9397 NMT2; N-myristoyltransferase 2 (EC:2.3.1.97); K00671
glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=498
Score = 144 bits (363), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 69/139 (49%), Positives = 92/139 (66%), Gaps = 6/139 (4%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K E G I+ K ++V++EPY+LP GF+W + D +VL EL
Sbjct 121 FWDTQPVPKLDE----VITSHGAIEPDK--DNVRQEPYSLPQGFMWDTLDLSDAEVLKEL 174
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
Y LL +YVEDDDN+FRF+Y +FL+WAL PP + +W GVRV +++KLVGFISAIP
Sbjct 175 YTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWLLQWHCGVRVSSNKKLVGFISAIPAN 234
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ ++ E+NFLCVH
Sbjct 235 IRIYDSVKKMVEINFLCVH 253
> mmu:18108 Nmt2, A930001K02Rik, AI605445, AU044698, hNMT-2; N-myristoyltransferase
2 (EC:2.3.1.97); K00671 glycylpeptide
N-tetradecanoyltransferase [EC:2.3.1.97]
Length=529
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 68/139 (48%), Positives = 93/139 (66%), Gaps = 6/139 (4%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K +E G I+ K +++++EPY+LP GF+W + + +VL EL
Sbjct 152 FWDTQPVPKLNE----VITSHGAIEPDK--DNIRQEPYSLPQGFMWDTLDLSNAEVLKEL 205
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
Y LL +YVEDDDN+FRF+Y +FL+WAL PP + +W GVRV +++KLVGFISAIP
Sbjct 206 YTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWLLQWHCGVRVSSNKKLVGFISAIPAN 265
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ R+ E+NFLCVH
Sbjct 266 IRIYDSVKRMVEINFLCVH 284
> dre:323956 nmt1a, im:2601337, nmt1, wu:fc15d01, wu:fc18a04,
zgc:110714; N-myristoyltransferase 1a (EC:2.3.1.97); K00671
glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=487
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 73/164 (44%), Positives = 100/164 (60%), Gaps = 13/164 (7%)
Query 6 QAIRGLSLSRSPLNS-------PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKK 58
+AI S+ + P + + FWDTQPV K E T G I+ K +++++
Sbjct 85 KAIELFSVGQGPAKTMEEATRRSYQFWDTQPVPKLGE----TVTSHGCIEPDK--DNIRE 138
Query 59 EPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
EPY+LP GF W + + VL ELY LL +YVEDDDN+FRF+Y +FL+WAL PP +
Sbjct 139 EPYSLPQGFTWDTLDLGNAAVLKELYTLLNENYVEDDDNMFRFDYSPEFLLWALRPPGWL 198
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
+W GVRV ++QKLVGFISAIP + ++ E+NFLCVH
Sbjct 199 PQWHCGVRVNSNQKLVGFISAIPANIRIYDIEKKMVEINFLCVH 242
> bbo:BBOV_III010870 17.m07935; myristoyl-CoA:protein N-myristoyltransferase,
N-terminal domain containing protein; K00671
glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=440
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 63/141 (44%), Positives = 92/141 (65%), Gaps = 2/141 (1%)
Query 22 HLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLD 81
H+FW+TQPV++ SE +++ + GPIDAT +V + PY LP+ F W + + D L
Sbjct 57 HVFWNTQPVLRFSED--VSSDEIGPIDATSSVSKIPTRPYLLPDAFEWVDIDINDETHLT 114
Query 82 ELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIP 141
+LY LL +YVED + +FRF+Y FL WA+ PP + K W +GVRV +S++LVGFIS +
Sbjct 115 QLYTLLNENYVEDGECMFRFDYKPAFLQWAMTPPGYKKNWHVGVRVRSSKRLVGFISGVA 174
Query 142 TTLMCNSKRIRVAEVNFLCVH 162
+ ++ AE+NFLCVH
Sbjct 175 ANIKVLGTSLKAAEINFLCVH 195
> tpv:TP02_0057 N-myristoyltransferase; K00671 glycylpeptide N-tetradecanoyltransferase
[EC:2.3.1.97]
Length=458
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 71/164 (43%), Positives = 104/164 (63%), Gaps = 5/164 (3%)
Query 2 ASLVQAIRGLSLSRSPLNS---PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKK 58
+S ++A S SP NS H FWDTQ V K ++ + + + GPID + V VKK
Sbjct 54 SSSIRARIFRRFSISPTNSYIPEHKFWDTQLVTKLTD--VVNSNECGPIDPNEDVSRVKK 111
Query 59 EPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
P LPNGF W + D + +++Y LL+ +YVED D LFRF+Y ++FL+WAL PN+
Sbjct 112 NPIPLPNGFEWISLDINDEEDRNQVYKLLSENYVEDGDALFRFDYKREFLIWALTVPNYN 171
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLCVH 162
K+W IGVRV + + L+G+I+A+P + +++AEVNFLC+H
Sbjct 172 KDWQIGVRVSSCKTLIGYITAVPVNVNVVGNTLKLAEVNFLCIH 215
> cel:T17E9.2 nmt-1; N-Myristoyl Transferase homolog family member
(nmt-1); K00671 glycylpeptide N-tetradecanoyltransferase
[EC:2.3.1.97]
Length=450
Score = 134 bits (336), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/139 (44%), Positives = 88/139 (63%), Gaps = 2/139 (1%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FW TQPV + E ++ I+ ++ V+ EP++LP GF WS + D + L+EL
Sbjct 68 FWSTQPVPQMDE--TVPADVNCAIEENIALDKVRAEPFSLPAGFRWSNVDLSDEEQLNEL 125
Query 84 YNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIPTT 143
YNLL +YVEDDD++FRF+Y DFL WAL P F EW GVR +++ +L+ FI A+P T
Sbjct 126 YNLLTRNYVEDDDSMFRFDYSADFLKWALQVPGFRPEWHCGVRADSNNRLLAFIGAVPQT 185
Query 144 LMCNSKRIRVAEVNFLCVH 162
+ K + + E+NFLCVH
Sbjct 186 VRVYDKTVNMVEINFLCVH 204
> sce:YLR195C NMT1, CDC72; N-myristoyl transferase, catalyzes
the cotranslational, covalent attachment of myristic acid to
the N-terminal glycine residue of several proteins involved
in cellular growth and signal transduction (EC:2.3.1.97); K00671
glycylpeptide N-tetradecanoyltransferase [EC:2.3.1.97]
Length=455
Score = 132 bits (331), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 68/141 (48%), Positives = 89/141 (63%), Gaps = 4/141 (2%)
Query 22 HLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLD 81
H FW TQPV E+ +EGPID KT ED+ +P L + F W V++ + L+
Sbjct 38 HKFWRTQPVKDFDEK----VVEEGPIDKPKTPEDISDKPLPLLSSFEWCSIDVDNKKQLE 93
Query 82 ELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIP 141
+++ LL +YVED D FRFNY K+F WAL P + K+W IGVRV+ +QKLV FISAIP
Sbjct 94 DVFVLLNENYVEDRDAGFRFNYTKEFFNWALKSPGWKKDWHIGVRVKETQKLVAFISAIP 153
Query 142 TTLMCNSKRIRVAEVNFLCVH 162
TL K++ E+NFLCVH
Sbjct 154 VTLGVRGKQVPSVEINFLCVH 174
> cpv:cgd3_320 N-myristoyltransferase ; K00671 glycylpeptide N-tetradecanoyltransferase
[EC:2.3.1.97]
Length=469
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 68/144 (47%), Positives = 92/144 (63%), Gaps = 6/144 (4%)
Query 21 PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVED--PQ 78
PH FW+TQPVV+ + S GPI+ + +KE Y LP+GF W +C++ D Q
Sbjct 44 PHKFWNTQPVVQNDDSSS--EYSFGPIEIEP--DSFRKEIYKLPDGFSWFDCNLWDIESQ 99
Query 79 VLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFIS 138
++ Y LL HYVEDDD+ FRFNY K+FL WAL P K W++GVRV ++K+VGFIS
Sbjct 100 DFEDTYQLLKDHYVEDDDSQFRFNYSKEFLRWALCVPGQKKNWLVGVRVNETKKMVGFIS 159
Query 139 AIPTTLMCNSKRIRVAEVNFLCVH 162
AIP + ++ + + VNFLCVH
Sbjct 160 AIPIKVRIHNCIMNTSVVNFLCVH 183
> dre:562651 nmt1b; si:dkeyp-94b4.2 (EC:2.3.1.97)
Length=471
Score = 111 bits (278), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 62/141 (43%), Positives = 87/141 (61%), Gaps = 8/141 (5%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWDTQPV K E+ + GPI++ V++E Y+LP GFVW ++ + L EL
Sbjct 92 FWDTQPVPKIDEKVTSC----GPIESE--WPKVREESYSLPQGFVWDTLNLTNQDQLREL 145
Query 84 YNLLALHYVEDD--DNLFRFNYGKDFLVWALNPPNFFKEWIIGVRVEASQKLVGFISAIP 141
+ L +Y+E++ DN R +Y FL+WAL PP + W GVRV+++QKLVGFISAIP
Sbjct 146 TDFLNENYMEEEVEDNRPRPHYSPQFLLWALCPPGWQSCWHCGVRVDSNQKLVGFISAIP 205
Query 142 TTLMCNSKRIRVAEVNFLCVH 162
T+ +++ E NFLCVH
Sbjct 206 ATIKTFDIELKMVEANFLCVH 226
> ath:AT2G44175 N-myristoyltransferase-related
Length=115
Score = 36.2 bits (82), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 34/70 (48%), Gaps = 2/70 (2%)
Query 27 TQPVVKASEQKSL--TTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDELY 84
T +VKA K + T+ E P++ + ++K+EP LP G+ W C + + E+
Sbjct 27 TDQIVKAKAHKDVGNTSLPESPVEPANPLSEIKQEPEKLPCGYEWITCDLNTDDMCSEVC 86
Query 85 NLLALHYVED 94
L Y+ D
Sbjct 87 KFLKEQYLVD 96
> ath:AT3G28250 glycosyl hydrolase family protein 17
Length=121
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 45/102 (44%), Gaps = 18/102 (17%)
Query 72 CSVEDPQVL---------DELYNL----LALHYVEDDDNLFRFNYGKDFLVWALNPPNFF 118
CS+ D Q++ D +YN+ + L+Y + N + N+G LV +P F+
Sbjct 23 CSIVDCQIISTRGACYSPDNIYNMASVVMNLYYQAEGRNFWNCNFGDSGLVAITDPSEFY 82
Query 119 KEWIIGVRVEASQKLVGFISAIPTTLMCNSKRIRVAEVNFLC 160
+ + + V F I + C + + VA +NF+C
Sbjct 83 ----LSLLFHYTIIYVLFFCLI-SNYFCFRQAMEVANMNFVC 119
> mmu:110596 Rgnef, 9230110L08Rik, AI323540, D13Bwg1089e, RIP2,
RhoGEF, Rhoip2, p190RhoGEF; Rho-guanine nucleotide exchange
factor
Length=1700
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 7/75 (9%)
Query 24 FWDTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFVWSECSVEDPQVLDEL 83
FWD +VKA EQ++ T + P A +T E+V+ NL +G SE ED + +
Sbjct 272 FWDRAFLVKALEQEAKTEKATMPSGAAETEEEVR----NLESGRSPSE-EEEDAKSIKSQ 326
Query 84 YNLLALHYVEDDDNL 98
+ + H ED D L
Sbjct 327 VDGPSEH--EDQDRL 339
> sce:YCL024W KCC4; Kcc4p (EC:2.7.11.1); K08286 protein-serine/threonine
kinase [EC:2.7.11.-]
Length=1037
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 45/96 (46%), Gaps = 14/96 (14%)
Query 21 PHLFWDTQPVVKASEQKSLTTQDEGPIDATKTVED-----VKKEPYNLPNGFVWSECSVE 75
PH ++ A+ ++S+ + + A +++D +K++P P G +E E
Sbjct 712 PHFTRKSKHFTTANNRRSVLS-----LYAKDSIKDLNEFLIKEDPDLPPQGSTDNESRSE 766
Query 76 DPQVLDELYNLLALHYVEDD----DNLFRFNYGKDF 107
DP++ + + + + Y EDD DN+ N DF
Sbjct 767 DPEIAESITDSRNIQYDEDDSKDGDNVNNDNILSDF 802
> mmu:67266 Fam69a, 2900024C23Rik, AI315274, MGC144843; family
with sequence similarity 69, member A
Length=428
Score = 29.3 bits (64), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 57/141 (40%), Gaps = 22/141 (15%)
Query 10 GLSLSRSPLNSPHL-FWDTQP-VVKASEQKSLTTQDEGPIDATK------------TVED 55
G LS P N +L WD P VVK +++L ++ K TV+
Sbjct 89 GKCLSNKPSNQMYLGVWDNLPGVVKCQMEQALHLDFGTELEPRKEIVLFDKPTRGTTVQK 148
Query 56 VKKEPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPP 115
K+ Y+L + D L EL NL+ V D D + + G+ WAL
Sbjct 149 FKEMVYSLFKA------KLGDQGNLSELVNLILT--VADGDRDGQVSLGEAKSAWALLQL 200
Query 116 NFFKEWIIGVRVEASQKLVGF 136
N F +I E + KL+GF
Sbjct 201 NEFLLMVILQDKEHTPKLMGF 221
> mmu:20129 Rptn; repetin
Length=1118
Score = 29.3 bits (64), Expect = 5.8, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query 51 KTVEDVKKEPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLF 99
K + V++EPY+ + W S E +E Y H VED +NL+
Sbjct 924 KLLAQVQQEPYSYEE-YDWQSQSSEQDHCGEEEYQDWDRHSVEDQENLY 971
> tgo:TGME49_028070 hypothetical protein
Length=3891
Score = 29.3 bits (64), Expect = 6.1, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 34/73 (46%), Gaps = 5/73 (6%)
Query 11 LSLSRSPLNSPHLFW--DTQPVVKASEQKSLTTQDEGPIDATKTVEDVKKEPYNLPNGFV 68
L L SP+ SPHLF +P +E++ T D G + A+ + N N +
Sbjct 2916 LRLHASPVYSPHLFQVHSEKPSPNTNERQGDLTADGGSVKASAATS---RRNANSQNPTL 2972
Query 69 WSECSVEDPQVLD 81
+ S+ D +VL+
Sbjct 2973 VARLSLTDTEVLE 2985
> hsa:388650 FAM69A, FLJ23493; family with sequence similarity
69, member A
Length=428
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 57/141 (40%), Gaps = 22/141 (15%)
Query 10 GLSLSRSPLNSPHL-FWDTQP-VVKASEQKSLTTQDEGPIDATK------------TVED 55
G LS P N +L WD P VVK +++L ++ K TV+
Sbjct 89 GKCLSTKPNNQMYLGIWDNLPGVVKCQMEQALHLDFGTELEPRKEIVLFDKPTRGTTVQK 148
Query 56 VKKEPYNLPNGFVWSECSVEDPQVLDELYNLLALHYVEDDDNLFRFNYGKDFLVWALNPP 115
K+ Y+L + D L EL NL+ V D D + + G+ WAL
Sbjct 149 FKEMVYSLFKA------KLGDQGNLSELVNLILT--VADGDKDGQVSLGEAKSAWALLQL 200
Query 116 NFFKEWIIGVRVEASQKLVGF 136
N F +I E + KL+GF
Sbjct 201 NEFLLMVILQDKEHTPKLMGF 221
Lambda K H
0.319 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40