bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2012_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
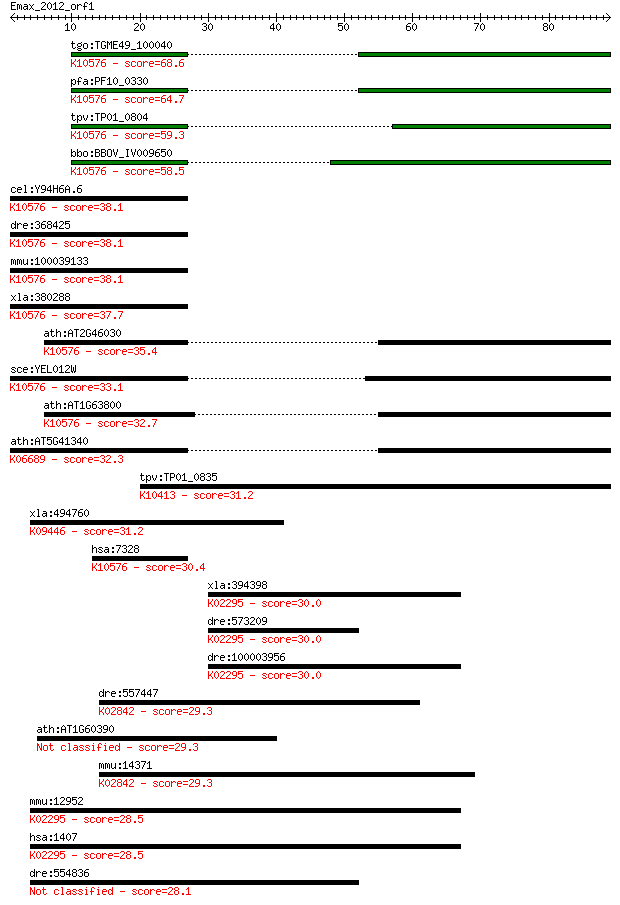
tgo:TGME49_100040 ubiquitin-conjugating enzyme E2, putative (E... 68.6 5e-12
pfa:PF10_0330 ubiquitin conjugating enzyme, putative; K10576 u... 64.7 6e-11
tpv:TP01_0804 ubiquitin-protein ligase; K10576 ubiquitin-conju... 59.3 3e-09
bbo:BBOV_IV009650 23.m05904; ubiquitin-conjugating enzyme subu... 58.5 5e-09
cel:Y94H6A.6 ubc-8; UBiquitin Conjugating enzyme family member... 38.1 0.007
dre:368425 ube2h, fc07a05, fi24c01, si:bz3c13.2, wu:fc07a05, w... 38.1 0.007
mmu:100039133 Gm2058; predicted gene 2058; K10576 ubiquitin-co... 38.1 0.007
xla:380288 ube2h, MGC53494; ubiquitin-conjugating enzyme E2H (... 37.7 0.008
ath:AT2G46030 UBC6; UBC6 (ubiquitin-conjugating enzyme 6); ubi... 35.4 0.050
sce:YEL012W UBC8, GID3; Ubc8p (EC:6.3.2.19); K10576 ubiquitin-... 33.1 0.21
ath:AT1G63800 UBC5; UBC5 (ubiquitin-conjugating enzyme 5); ubi... 32.7 0.30
ath:AT5G41340 UBC4; UBC4 (UBIQUITIN CONJUGATING ENZYME 4); ubi... 32.3 0.39
tpv:TP01_0835 hypothetical protein; K10413 dynein heavy chain ... 31.2 0.73
xla:494760 irf5; interferon regulatory factor 5; K09446 interf... 31.2 0.88
hsa:7328 UBE2H, E2-20K, UBC8, UBCH, UBCH2; ubiquitin-conjugati... 30.4 1.4
xla:394398 cry1, cry1-A, xCRY1; cryptochrome 1 (photolyase-lik... 30.0 1.8
dre:573209 cry2a, MGC110521, zgc:110521; cryptochrome 2a; K022... 30.0 1.9
dre:100003956 cry1a, MGC153885, zcry1a, zgc:153885; cryptochro... 30.0 1.9
dre:557447 fzd9; frizzled homolog 9 (Drosophila); K02842 frizz... 29.3 3.0
ath:AT1G60390 BURP domain-containing protein / polygalacturona... 29.3 3.2
mmu:14371 Fzd9, mfz9; frizzled homolog 9 (Drosophila); K02842 ... 29.3 3.5
mmu:12952 Cry1, AU020726, AU021000, Phll1; cryptochrome 1 (pho... 28.5 5.5
hsa:1407 CRY1, PHLL1; cryptochrome 1 (photolyase-like); K02295... 28.5 6.1
dre:554836 cry1b, fi26e03, wu:fi26e03, zcry1b; cryptochrome 1b 28.1 6.8
> tgo:TGME49_100040 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10576 ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=240
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 29/37 (78%), Positives = 35/37 (94%), Gaps = 0/37 (0%)
Query 52 QISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
Q+SS+RKQCDF KL+MAG+DLELNN +TQDF+VVFHG
Sbjct 13 QVSSNRKQCDFTKLMMAGFDLELNNDSTQDFHVVFHG 49
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/17 (88%), Positives = 17/17 (100%), Gaps = 0/17 (0%)
Query 10 IGFLNKILHPNVDEASG 26
IGF+NK+LHPNVDEASG
Sbjct 77 IGFMNKMLHPNVDEASG 93
> pfa:PF10_0330 ubiquitin conjugating enzyme, putative; K10576
ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=191
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 28/37 (75%), Positives = 33/37 (89%), Gaps = 0/37 (0%)
Query 52 QISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
Q S +RKQCDF KL+MAGYDLELNN +TQDF+V+FHG
Sbjct 4 QTSLTRKQCDFTKLIMAGYDLELNNGSTQDFDVMFHG 40
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 15/17 (88%), Positives = 17/17 (100%), Gaps = 0/17 (0%)
Query 10 IGFLNKILHPNVDEASG 26
IGF+NK+LHPNVDEASG
Sbjct 68 IGFMNKLLHPNVDEASG 84
> tpv:TP01_0804 ubiquitin-protein ligase; K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=189
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 26/32 (81%), Positives = 28/32 (87%), Gaps = 0/32 (0%)
Query 57 RKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
RKQCDF KLLMAGYDLEL N +TQ+FNV FHG
Sbjct 9 RKQCDFTKLLMAGYDLELVNGSTQEFNVTFHG 40
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 14/17 (82%), Positives = 17/17 (100%), Gaps = 0/17 (0%)
Query 10 IGFLNKILHPNVDEASG 26
IGF+NK+LHPNVDE+SG
Sbjct 68 IGFMNKMLHPNVDESSG 84
> bbo:BBOV_IV009650 23.m05904; ubiquitin-conjugating enzyme subunit
(EC:6.3.2.19); K10576 ubiquitin-conjugating enzyme E2
H [EC:6.3.2.19]
Length=191
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/41 (68%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 48 MTQVQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
M V IS+ RKQ DF KLLMAGYDLEL N N +FNV FHG
Sbjct 1 MNNVSISNVRKQNDFTKLLMAGYDLELINGNMTEFNVTFHG 41
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 15/17 (88%), Positives = 16/17 (94%), Gaps = 0/17 (0%)
Query 10 IGFLNKILHPNVDEASG 26
IGFLNK+LHPNVDE SG
Sbjct 69 IGFLNKMLHPNVDETSG 85
> cel:Y94H6A.6 ubc-8; UBiquitin Conjugating enzyme family member
(ubc-8); K10576 ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=221
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 17/26 (65%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 1 DRRRRKRIIIGFLNKILHPNVDEASG 26
D+ K IGFLNKI HPN+DEASG
Sbjct 60 DKYPFKSPSIGFLNKIFHPNIDEASG 85
> dre:368425 ube2h, fc07a05, fi24c01, si:bz3c13.2, wu:fc07a05,
wu:fi24c01; ubiquitin-conjugating enzyme E2H (UBC8 homolog,
yeast) (EC:6.3.2.19); K10576 ubiquitin-conjugating enzyme E2
H [EC:6.3.2.19]
Length=183
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/26 (61%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 1 DRRRRKRIIIGFLNKILHPNVDEASG 26
D+ K IGF+NKI HPN+DEASG
Sbjct 59 DKYPFKSPSIGFMNKIFHPNIDEASG 84
> mmu:100039133 Gm2058; predicted gene 2058; K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=183
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/26 (61%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 1 DRRRRKRIIIGFLNKILHPNVDEASG 26
D+ K IGF+NKI HPN+DEASG
Sbjct 59 DKYPFKSPSIGFMNKIFHPNIDEASG 84
> xla:380288 ube2h, MGC53494; ubiquitin-conjugating enzyme E2H
(UBC8 homolog) (EC:6.3.2.19); K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=183
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 16/26 (61%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 1 DRRRRKRIIIGFLNKILHPNVDEASG 26
D+ K IGF+NKI HPN+DEASG
Sbjct 59 DKYPFKSPSIGFMNKIFHPNIDEASG 84
> ath:AT2G46030 UBC6; UBC6 (ubiquitin-conjugating enzyme 6); ubiquitin-protein
ligase; K10576 ubiquitin-conjugating enzyme
E2 H [EC:6.3.2.19]
Length=183
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 14/21 (66%), Positives = 17/21 (80%), Gaps = 0/21 (0%)
Query 6 KRIIIGFLNKILHPNVDEASG 26
K +GF+NKI HPNVDE+SG
Sbjct 62 KSPSVGFVNKIYHPNVDESSG 82
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 55 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
S R++ D KL+M+ Y ++ N + Q F V FHG
Sbjct 5 SKRREMDMMKLMMSDYKVDTVNDDLQMFYVTFHG 38
> sce:YEL012W UBC8, GID3; Ubc8p (EC:6.3.2.19); K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=218
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 1 DRRRRKRIIIGFLNKILHPNVDEASG 26
D K IGF+NKI HPN+D ASG
Sbjct 57 DNYPYKSPSIGFVNKIFHPNIDIASG 82
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 26/38 (68%), Gaps = 2/38 (5%)
Query 53 ISSSRK--QCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
+SSS++ + D KLLM+ + ++L N + Q+F+V F G
Sbjct 1 MSSSKRRIETDVMKLLMSDHQVDLINDSMQEFHVKFLG 38
> ath:AT1G63800 UBC5; UBC5 (ubiquitin-conjugating enzyme 5); ubiquitin-protein
ligase; K10576 ubiquitin-conjugating enzyme
E2 H [EC:6.3.2.19]
Length=185
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 6 KRIIIGFLNKILHPNVDEASGH 27
K +GF+ KI HPNVDE SG
Sbjct 62 KSPSVGFITKIYHPNVDEMSGS 83
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 55 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
S R++ D KL+M+ Y +E+ N Q+F V F G
Sbjct 5 SKRREMDLMKLMMSDYKVEMINDGMQEFFVEFSG 38
> ath:AT5G41340 UBC4; UBC4 (UBIQUITIN CONJUGATING ENZYME 4); ubiquitin-protein
ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=187
Score = 32.3 bits (72), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 14/26 (53%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 1 DRRRRKRIIIGFLNKILHPNVDEASG 26
D K +GF+ KI HPNVDE SG
Sbjct 57 DAYPYKSPSVGFITKIYHPNVDELSG 82
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 55 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHG 88
S R++ D KL+M+ Y +E N Q+F V F+G
Sbjct 5 SKRREMDMMKLMMSDYKVETINDGMQEFYVEFNG 38
> tpv:TP01_0835 hypothetical protein; K10413 dynein heavy chain
1, cytosolic
Length=1970
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 38/84 (45%), Gaps = 15/84 (17%)
Query 20 NVDEASGHFQGLPPYLRFWCFLCQIYLKMTQVQISSSRKQCDFAKL------------LM 67
NV+ +S F G+P L F +C +Y ++ + C F L L+
Sbjct 8 NVNLSSKVFTGIPDSLSFISCICDLYFNVSDASVIDILNNCYFDTLSNFIDQNNPEIPLI 67
Query 68 AGYDLELNNKNTQDF-NVVF--HG 88
G+ +L+++N +F N VF HG
Sbjct 68 IGFKDDLDSENAHEFSNRVFVSHG 91
> xla:494760 irf5; interferon regulatory factor 5; K09446 interferon
regulatory factor 5
Length=517
Score = 31.2 bits (69), Expect = 0.88, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 4 RRKRIIIGFLNKILHPNVDEASGHFQGLPPYLRFWCF 40
R KRI + L L+ + GH LPPY F+CF
Sbjct 381 REKRIKLFSLETFLNELIACQKGHTSSLPPYEIFFCF 417
> hsa:7328 UBE2H, E2-20K, UBC8, UBCH, UBCH2; ubiquitin-conjugating
enzyme E2H (UBC8 homolog, yeast) (EC:6.3.2.19); K10576
ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=113
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 11/14 (78%), Positives = 13/14 (92%), Gaps = 0/14 (0%)
Query 13 LNKILHPNVDEASG 26
+NKI HPN+DEASG
Sbjct 1 MNKIFHPNIDEASG 14
> xla:394398 cry1, cry1-A, xCRY1; cryptochrome 1 (photolyase-like);
K02295 cryptochrome
Length=616
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 5/42 (11%)
Query 30 GLPPYLRFWCFLCQI-YLKMT----QVQISSSRKQCDFAKLL 66
GL PYLRF C C++ Y K+T +V+ +SS + +LL
Sbjct 250 GLSPYLRFGCLSCRLFYFKLTDLYKKVKKNSSPPLSLYGQLL 291
> dre:573209 cry2a, MGC110521, zgc:110521; cryptochrome 2a; K02295
cryptochrome
Length=655
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 1/23 (4%)
Query 30 GLPPYLRFWCFLCQI-YLKMTQV 51
GL PYLRF C C++ Y K+T +
Sbjct 250 GLSPYLRFGCLSCRLFYFKLTDL 272
> dre:100003956 cry1a, MGC153885, zcry1a, zgc:153885; cryptochrome
1a; K02295 cryptochrome
Length=619
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 5/42 (11%)
Query 30 GLPPYLRFWCFLCQI-YLKMT----QVQISSSRKQCDFAKLL 66
GL PYLRF C C++ Y K+T +V+ +SS + +LL
Sbjct 250 GLSPYLRFGCLSCRLFYFKLTDLYRKVKKNSSPPLSLYGQLL 291
> dre:557447 fzd9; frizzled homolog 9 (Drosophila); K02842 frizzled
9/10
Length=581
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 26/53 (49%), Gaps = 6/53 (11%)
Query 14 NKILHPNVDEASGHFQGLPPYLRFWC------FLCQIYLKMTQVQISSSRKQC 60
N + + + +EAS P +++ C FLC +Y+ M Q+S+S C
Sbjct 56 NLLNYESQEEASIKLNEFAPLVKYGCDIHLQFFLCSLYVPMCAEQVSASIPAC 108
> ath:AT1G60390 BURP domain-containing protein / polygalacturonase,
putative
Length=624
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 5 RKRIIIGFLNKILHPNVDEASGHFQGLPPYLRFWC 39
+K+++IG +N I +V A Q L PYL ++C
Sbjct 520 KKKVVIGKVNGINGGDVTRAVSCHQSLYPYLLYYC 554
> mmu:14371 Fzd9, mfz9; frizzled homolog 9 (Drosophila); K02842
frizzled 9/10
Length=592
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 29/65 (44%), Gaps = 10/65 (15%)
Query 14 NKILHPNVDEASGHFQGLPPYLRFWC------FLCQIYLKMTQVQISSS----RKQCDFA 63
N + H + EA+ P +++ C FLC +Y M Q+S+ R C+ A
Sbjct 60 NLLGHTSQGEAAAQLAEFSPLVQYGCHSHLRFFLCSLYAPMCTDQVSTPIPACRPMCEQA 119
Query 64 KLLMA 68
+L A
Sbjct 120 RLRCA 124
> mmu:12952 Cry1, AU020726, AU021000, Phll1; cryptochrome 1 (photolyase-like);
K02295 cryptochrome
Length=606
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 34/68 (50%), Gaps = 7/68 (10%)
Query 4 RRKRIIIGFLNKILHPNVDEASGHFQGLPPYLRFWCFLCQI-YLKMT----QVQISSSRK 58
RK + F ++ N AS GL PYLRF C C++ Y K+T +V+ +SS
Sbjct 226 ERKAWVANFERPRMNANSLLASP--TGLSPYLRFGCLSCRLFYFKLTDLYKKVKKNSSPP 283
Query 59 QCDFAKLL 66
+ +LL
Sbjct 284 LSLYGQLL 291
> hsa:1407 CRY1, PHLL1; cryptochrome 1 (photolyase-like); K02295
cryptochrome
Length=586
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 34/68 (50%), Gaps = 7/68 (10%)
Query 4 RRKRIIIGFLNKILHPNVDEASGHFQGLPPYLRFWCFLCQI-YLKMT----QVQISSSRK 58
RK + F ++ N AS GL PYLRF C C++ Y K+T +V+ +SS
Sbjct 226 ERKAWVANFERPRMNANSLLASP--TGLSPYLRFGCLSCRLFYFKLTDLYKKVKKNSSPP 283
Query 59 QCDFAKLL 66
+ +LL
Sbjct 284 LSLYGQLL 291
> dre:554836 cry1b, fi26e03, wu:fi26e03, zcry1b; cryptochrome
1b
Length=606
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 25/49 (51%), Gaps = 3/49 (6%)
Query 4 RRKRIIIGFLNKILHPNVDEASGHFQGLPPYLRFWCFLCQI-YLKMTQV 51
RK + F ++ N AS GL PYLRF C C++ Y K+T +
Sbjct 226 ERKAWVANFERPRMNANSLLASP--TGLSPYLRFGCLSCRLFYFKLTDL 272
Lambda K H
0.329 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2021645584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40