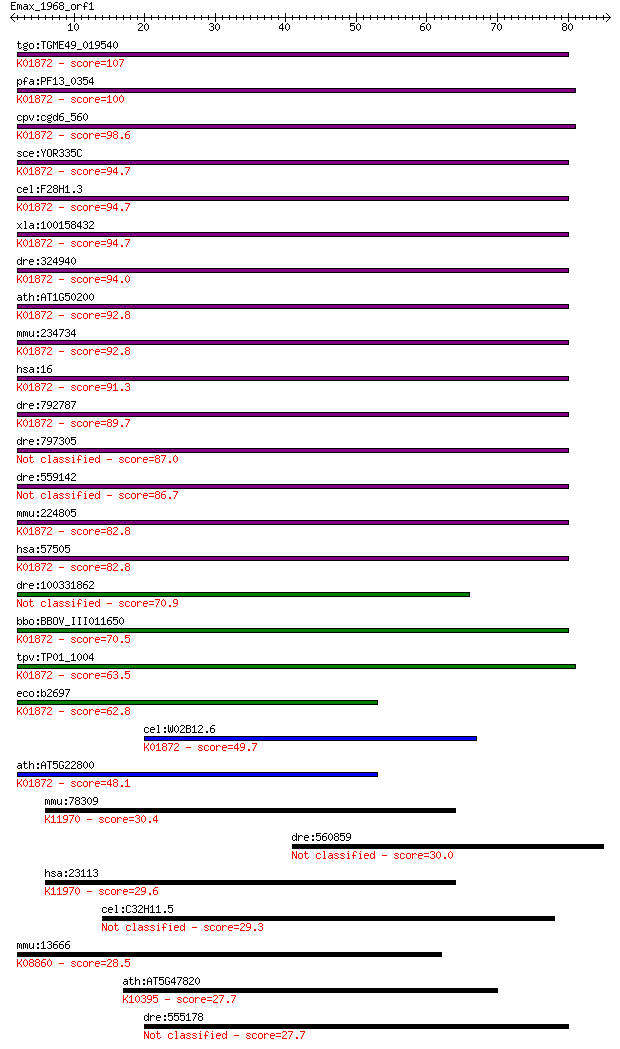
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1968_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019540 alanyl-tRNA synthetase, putative (EC:6.1.1.7... 107 1e-23
pfa:PF13_0354 alanine--tRNA ligase, putative (EC:6.1.1.7); K01... 100 1e-21
cpv:cgd6_560 alanyl-tRNA synthetase (with HxxxH domain) ; K018... 98.6 5e-21
sce:YOR335C ALA1, CDC64; Ala1p (EC:6.1.1.7); K01872 alanyl-tRN... 94.7 6e-20
cel:F28H1.3 ars-2; Alanyl tRNA Synthetase family member (ars-2... 94.7 6e-20
xla:100158432 aars; alanyl-tRNA synthetase (EC:6.1.1.7); K0187... 94.7 7e-20
dre:324940 aars, MGC113920, MGC158129, MGC198394, MGC198396, i... 94.0 1e-19
ath:AT1G50200 ALATS; ALATS (ALANYL-TRNA SYNTHETASE); ATP bindi... 92.8 2e-19
mmu:234734 Aars, AI316495, C76919, MGC37368, sti; alanyl-tRNA ... 92.8 2e-19
hsa:16 AARS, CMT2N; alanyl-tRNA synthetase (EC:6.1.1.7); K0187... 91.3 7e-19
dre:792787 im:7141191; si:dkey-240e12.1; K01872 alanyl-tRNA sy... 89.7 2e-18
dre:797305 Alanyl tRNA Synthetase family member (ars-2)-like 87.0 1e-17
dre:559142 Alanyl tRNA Synthetase family member (ars-2)-like 86.7 2e-17
mmu:224805 Aars2, Aarsl, AlaRS, Gm89, MGC69820; alanyl-tRNA sy... 82.8 2e-16
hsa:57505 AARS2, AARSL, KIAA1270, MT-ALARS, MTALARS, bA444E17.... 82.8 2e-16
dre:100331862 Alanyl tRNA Synthetase family member (ars-2)-like 70.9 9e-13
bbo:BBOV_III011650 17.m07993; alanyl-tRNA synthetase family pr... 70.5 1e-12
tpv:TP01_1004 alanyl-tRNA synthetase; K01872 alanyl-tRNA synth... 63.5 2e-10
eco:b2697 alaS, act, ECK2692, JW2667, lovB; alanyl-tRNA synthe... 62.8 2e-10
cel:W02B12.6 ars-1; Alanyl tRNA Synthetase family member (ars-... 49.7 2e-06
ath:AT5G22800 EMB1030 (EMBRYO DEFECTIVE 1030); ATP binding / a... 48.1 7e-06
mmu:78309 Cul9, 1810035I07Rik, Parc, mKIAA0708; cullin 9; K119... 30.4 1.4
dre:560859 hypothetical LOC560859 30.0 2.1
hsa:23113 CUL9, DKFZp686G1042, DKFZp686P2024, H7AP1, PARC, RP3... 29.6 2.7
cel:C32H11.5 hypothetical protein 29.3 3.6
mmu:13666 Eif2ak3, AI427929, Pek, Perk; eukaryotic translation... 28.5 5.4
ath:AT5G47820 FRA1; FRA1 (FRAGILE FIBER 1); microtubule motor;... 27.7 8.7
dre:555178 ifrd1, MGC154080, im:7067566, si:dkey-192l17.2, wu:... 27.7 8.8
> tgo:TGME49_019540 alanyl-tRNA synthetase, putative (EC:6.1.1.7);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1037
Score = 107 bits (266), Expect = 1e-23, Method: Composition-based stats.
Identities = 49/78 (62%), Positives = 60/78 (76%), Gaps = 0/78 (0%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ+NRE +LTPLPAPCVDTG LER+ SVLQ+K+SNYDTDLF +F++I + L
Sbjct 239 MQYNREKDGSLTPLPAPCVDTGMGLERVASVLQDKQSNYDTDLFTALFDKILELSPGLRP 298
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVGEEDK+ +D AYR
Sbjct 299 YGGKVGEEDKDMVDMAYR 316
> pfa:PF13_0354 alanine--tRNA ligase, putative (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1408
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 44/79 (55%), Positives = 58/79 (73%), Gaps = 0/79 (0%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ+N++ + + LP PC+DTG LER+ S+LQN SNYDTDLFQPIF++I LP
Sbjct 604 MQYNKDENKNMNKLPFPCIDTGMGLERITSILQNVDSNYDTDLFQPIFKQIKELFPYLPN 663
Query 62 YQGKVGEEDKNKIDTAYRA 80
Y+GK+ E+D +KIDTAYR
Sbjct 664 YEGKINEQDVDKIDTAYRV 682
> cpv:cgd6_560 alanyl-tRNA synthetase (with HxxxH domain) ; K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1003
Score = 98.6 bits (244), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 49/84 (58%), Positives = 60/84 (71%), Gaps = 5/84 (5%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIH----SFNT 57
MQ+ RE TLTPLP CVDTG LERLVS+LQNK SNYD DLF+PIF++IH S
Sbjct 236 MQYYREVDSTLTPLPKKCVDTGMGLERLVSILQNKTSNYDIDLFKPIFDQIHQVISSSGV 295
Query 58 KLPKYQGKVGE-EDKNKIDTAYRA 80
++P+Y GKVG+ +D ID +YR
Sbjct 296 QIPRYSGKVGDIDDPEMIDMSYRV 319
> sce:YOR335C ALA1, CDC64; Ala1p (EC:6.1.1.7); K01872 alanyl-tRNA
synthetase [EC:6.1.1.7]
Length=958
Score = 94.7 bits (234), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 55/78 (70%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE +L PLPA +DTG ERLVSVLQ+ +SNYDTD+F P+FERI + P
Sbjct 224 IQFNREQDGSLKPLPAKHIDTGMGFERLVSVLQDVRSNYDTDVFTPLFERIQEITSVRP- 282
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G GE DK+ IDTAYR
Sbjct 283 YSGNFGENDKDGIDTAYR 300
> cel:F28H1.3 ars-2; Alanyl tRNA Synthetase family member (ars-2);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 94.7 bits (234), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 43/78 (55%), Positives = 56/78 (71%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE L PLPA +D G LERL++V+Q+K SNYDTD+FQPIFE IH + +
Sbjct 220 IQFNREEGGVLKPLPAKHIDCGLGLERLIAVMQDKTSNYDTDIFQPIFEAIHK-GSGVRA 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G +G+EDK+ +D AYR
Sbjct 279 YTGFIGDEDKDGVDMAYR 296
> xla:100158432 aars; alanyl-tRNA synthetase (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 94.7 bits (234), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 49/78 (62%), Positives = 53/78 (67%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE TL PLP +DTG LERLVSVLQNK SNYDTDLF P FE I P
Sbjct 220 IQFNRETDGTLKPLPKKSIDTGMGLERLVSVLQNKMSNYDTDLFVPYFEAIQKGTGARP- 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG+ED + ID AYR
Sbjct 279 YTGKVGDEDVDGIDMAYR 296
> dre:324940 aars, MGC113920, MGC158129, MGC198394, MGC198396,
im:7146712, si:ch211-223o1.6, wu:fc48h07, zgc:113920; alanyl-tRNA
synthetase (EC:6.1.1.7); K01872 alanyl-tRNA synthetase
[EC:6.1.1.7]
Length=433
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 48/78 (61%), Positives = 52/78 (66%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE+ L PLP +DTG LERLVSVLQNK SNYDTDLF P FE I T
Sbjct 220 IQFNRESETVLKPLPKKSIDTGMGLERLVSVLQNKMSNYDTDLFVPYFEAIQK-GTGARA 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG ED + ID AYR
Sbjct 279 YTGKVGAEDTDGIDMAYR 296
> ath:AT1G50200 ALATS; ALATS (ALANYL-TRNA SYNTHETASE); ATP binding
/ alanine-tRNA ligase/ ligase, forming aminoacyl-tRNA and
related compounds / nucleic acid binding / nucleotide binding
(EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1003
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 54/78 (69%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE+ +L PLPA VDTG ERL SVLQNK SNYDTD+F PIF+ I P
Sbjct 273 IQFNRESDGSLKPLPAKHVDTGMGFERLTSVLQNKMSNYDTDVFMPIFDDIQKATGARP- 331
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG ED +++D AYR
Sbjct 332 YSGKVGPEDVDRVDMAYR 349
> mmu:234734 Aars, AI316495, C76919, MGC37368, sti; alanyl-tRNA
synthetase (EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 52/78 (66%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+Q+NRE+ L PLP +DTG LERLVSVLQNK SNYDTDLF P FE I P
Sbjct 220 IQYNRESDGVLKPLPKKSIDTGMGLERLVSVLQNKMSNYDTDLFMPYFEAIQKGTGARP- 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG ED + ID AYR
Sbjct 279 YTGKVGAEDADGIDMAYR 296
> hsa:16 AARS, CMT2N; alanyl-tRNA synthetase (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 91.3 bits (225), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 47/78 (60%), Positives = 51/78 (65%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+Q+NRE L PLP +DTG LERLVSVLQNK SNYDTDLF P FE I P
Sbjct 220 IQYNREADGILKPLPKKSIDTGMGLERLVSVLQNKMSNYDTDLFVPYFEAIQKGTGARP- 278
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKVG ED + ID AYR
Sbjct 279 YTGKVGAEDADGIDMAYR 296
> dre:792787 im:7141191; si:dkey-240e12.1; K01872 alanyl-tRNA
synthetase [EC:6.1.1.7]
Length=1004
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 43/78 (55%), Positives = 53/78 (67%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ+NRE+ +L LP VDTG LERLV+VLQ K+SNYDTDLF P+ IH +K P
Sbjct 258 MQYNRESNGSLRALPQCSVDTGMGLERLVTVLQGKRSNYDTDLFTPLLSAIHQC-SKAPA 316
Query 62 YQGKVGEEDKNKIDTAYR 79
YQG+ G D ++D AYR
Sbjct 317 YQGRTGAADVAQLDMAYR 334
> dre:797305 Alanyl tRNA Synthetase family member (ars-2)-like
Length=305
Score = 87.0 bits (214), Expect = 1e-17, Method: Composition-based stats.
Identities = 46/78 (58%), Positives = 50/78 (64%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE+ L PL +DTG LERLVSVLQNK SNYDTDLF P FE I T
Sbjct 115 IQFNRESETVLKPLSKKSIDTGMGLERLVSVLQNKMSNYDTDLFVPYFEAIQK-GTGARA 173
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKV ED + ID AYR
Sbjct 174 YTGKVSAEDTDGIDMAYR 191
> dre:559142 Alanyl tRNA Synthetase family member (ars-2)-like
Length=725
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 46/78 (58%), Positives = 50/78 (64%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
+QFNRE+ L PL +DTG LERLVSVLQNK SNYDTDLF P FE I T
Sbjct 105 IQFNRESETVLKPLSKKSIDTGMGLERLVSVLQNKMSNYDTDLFVPYFEAIQK-GTGARA 163
Query 62 YQGKVGEEDKNKIDTAYR 79
Y GKV ED + ID AYR
Sbjct 164 YTGKVSAEDTDGIDMAYR 181
> mmu:224805 Aars2, Aarsl, AlaRS, Gm89, MGC69820; alanyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=980
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 42/78 (53%), Positives = 51/78 (65%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ RE +L LP VDTG LERLV+VLQ K+S YDTDLF P+ + IH + P
Sbjct 241 MQHYREADGSLQLLPQRHVDTGMGLERLVAVLQGKRSTYDTDLFSPLLDAIHQ-SCGAPP 299
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G+VG D+ +IDTAYR
Sbjct 300 YSGRVGAADEGRIDTAYR 317
> hsa:57505 AARS2, AARSL, KIAA1270, MT-ALARS, MTALARS, bA444E17.1;
alanyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.7);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=985
Score = 82.8 bits (203), Expect = 2e-16, Method: Composition-based stats.
Identities = 42/78 (53%), Positives = 49/78 (62%), Gaps = 1/78 (1%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ NRE +L PLP VDTG LERLV+VLQ K S YDTDLF P+ I + P
Sbjct 246 MQHNREADGSLQPLPQRHVDTGMGLERLVAVLQGKHSTYDTDLFSPLLNAIQQ-GCRAPP 304
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G+VG D+ + DTAYR
Sbjct 305 YLGRVGVADEGRTDTAYR 322
> dre:100331862 Alanyl tRNA Synthetase family member (ars-2)-like
Length=796
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 33/64 (51%), Positives = 43/64 (67%), Gaps = 0/64 (0%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ+NRE+ +L LP VDTG LERLV+VLQ K+SNYDTDLF P+ IH + +
Sbjct 258 MQYNRESNGSLRALPQCSVDTGMGLERLVTVLQGKRSNYDTDLFTPLLSAIHQLGLQSEE 317
Query 62 YQGK 65
+G+
Sbjct 318 DEGE 321
> bbo:BBOV_III011650 17.m07993; alanyl-tRNA synthetase family
protein (EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=978
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 47/78 (60%), Gaps = 5/78 (6%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ+NR++ ++ LP PCVDTG LER+ +VL++ SNYD DLF IF ++ LP
Sbjct 235 MQYNRKSDGSVELLPRPCVDTGMGLERVCAVLKSSNSNYDGDLFTDIFAYVNKLMPHLPP 294
Query 62 YQGKVGEEDKNKIDTAYR 79
Y G + ID AYR
Sbjct 295 YGGS-----DSMIDVAYR 307
> tpv:TP01_1004 alanyl-tRNA synthetase; K01872 alanyl-tRNA synthetase
[EC:6.1.1.7]
Length=1011
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK 61
MQ+NR L LP CVDTG LERL SVL N SNYDTDL + I +L
Sbjct 271 MQYNRSRNGVLERLPKGCVDTGMGLERLCSVLNNTNSNYDTDLLNDVILYISELLPELES 330
Query 62 YQGKVGEEDKNKIDTAYRA 80
Y+G + +D AYR
Sbjct 331 YKGG-----NSPLDVAYRV 344
> eco:b2697 alaS, act, ECK2692, JW2667, lovB; alanyl-tRNA synthetase
(EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=876
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/51 (52%), Positives = 36/51 (70%), Gaps = 0/51 (0%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERI 52
MQFNR+ T+ PLP P VDTG LER+ +VLQ+ SNYD DLF+ + + +
Sbjct 217 MQFNRQADGTMEPLPKPSVDTGMGLERIAAVLQHVNSNYDIDLFRTLIQAV 267
> cel:W02B12.6 ars-1; Alanyl tRNA Synthetase family member (ars-1);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=793
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 2/47 (4%)
Query 20 VDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPKYQGKV 66
+DTG ERL+SV+QNK SN+DTD+F PI E+ K +Y G +
Sbjct 248 IDTGMGFERLLSVVQNKTSNFDTDVFTPILEKTSELAKK--QYTGSL 292
> ath:AT5G22800 EMB1030 (EMBRYO DEFECTIVE 1030); ATP binding /
alanine-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleic acid binding / nucleotide binding (EC:6.1.1.7);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=978
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 22/51 (43%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERI 52
MQ+N+ L PL +DTG LER+ +LQ +NY+TDL PI +I
Sbjct 277 MQYNKTEDGLLEPLKQKNIDTGLGLERIAQILQKVPNNYETDLIYPIIAKI 327
> mmu:78309 Cul9, 1810035I07Rik, Parc, mKIAA0708; cullin 9; K11970
p53-associated parkin-like cytoplasmic protein
Length=2520
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 31/63 (49%), Gaps = 5/63 (7%)
Query 6 RENAQTLTPLPAPCVDTGTVL-----ERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLP 60
R+ A +L L A C+ TGT L ERL+++LQ+ ++ L P E + +P
Sbjct 2395 RDLASSLRFLRADCLSTGTELLRRIQERLLAILQHSTQDFRVGLQSPSVETREVKGSNVP 2454
Query 61 KYQ 63
Q
Sbjct 2455 SDQ 2457
> dre:560859 hypothetical LOC560859
Length=408
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 7/51 (13%)
Query 41 DTDLFQPIFERIHSFNTKLPK----YQGKVGEEDKNKID---TAYRAGGQV 84
D D+FQP ++FN P+ QG +G ED+ +D ++A G +
Sbjct 326 DKDVFQPFLTPPNTFNLPRPRSPLISQGSLGFEDRRMVDNELNTFKAAGDI 376
> hsa:23113 CUL9, DKFZp686G1042, DKFZp686P2024, H7AP1, PARC, RP3-330M21.2;
cullin 9; K11970 p53-associated parkin-like cytoplasmic
protein
Length=2517
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 5/63 (7%)
Query 6 RENAQTLTPLPAPCVDTGTVL-----ERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLP 60
R+ A +L L A C+ TG L ERL+++LQ+ ++ L P E + +P
Sbjct 2391 RDLASSLRLLRADCLSTGMELLRRIQERLLAILQHSAQDFRVGLQSPSVEAWEAKGPNMP 2450
Query 61 KYQ 63
Q
Sbjct 2451 GSQ 2453
> cel:C32H11.5 hypothetical protein
Length=194
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 6/67 (8%)
Query 14 PLPAPCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFER---IHSFNTKLPKYQGKVGEED 70
P PAP T TV E + ++ QN D FQ +F+ + + K ++ K G D
Sbjct 19 PTPAP---TLTVAEEMEAIFQNLTEEAQNDYFQILFDHTLTLAELDQKWDEWAEKHGIAD 75
Query 71 KNKIDTA 77
K + T+
Sbjct 76 KWAVYTS 82
> mmu:13666 Eif2ak3, AI427929, Pek, Perk; eukaryotic translation
initiation factor 2 alpha kinase 3 (EC:2.7.11.1); K08860
eukaryotic translation initiation factor 2-alpha kinase [EC:2.7.11.1]
Length=1114
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 2 MQFNRENAQTLTPLPAPCVDTGTVLERL-VSVLQNKKSNYD--TDLFQ---PIFERIHSF 55
M + E LTP+PA TG V +L +S Q +NY D+F +FE ++ F
Sbjct 960 MDQDEEEQTVLTPMPAYATHTGQVGTKLYMSPEQIHGNNYSHKVDIFSLGLILFELLYPF 1019
Query 56 NTKLPK 61
+T++ +
Sbjct 1020 STQMER 1025
> ath:AT5G47820 FRA1; FRA1 (FRAGILE FIBER 1); microtubule motor;
K10395 kinesin family member 4/7/21/27
Length=1035
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 3/56 (5%)
Query 17 APCVDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPK---YQGKVGEE 69
A C+D G+V S N +S+ +F E++ NT P+ Y G + EE
Sbjct 206 AACLDQGSVSRATGSTNMNNQSSRSHAIFTITVEQMRKINTDSPENGAYNGSLKEE 261
> dre:555178 ifrd1, MGC154080, im:7067566, si:dkey-192l17.2, wu:fi35f04,
wu:fj67a06, zgc:154080; interferon-related developmental
regulator 1
Length=427
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 13/60 (21%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 20 VDTGTVLERLVSVLQNKKSNYDTDLFQPIFERIHSFNTKLPKYQGKVGEEDKNKIDTAYR 79
+ G + L ++++ + +D D ++P+ E++++ T K++ K DK K + +R
Sbjct 261 IAAGETIALLFELIRDVEPEFDLDEWEPLCEKLNALATDCNKHRAKT---DKRKQRSVFR 317
Lambda K H
0.317 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40