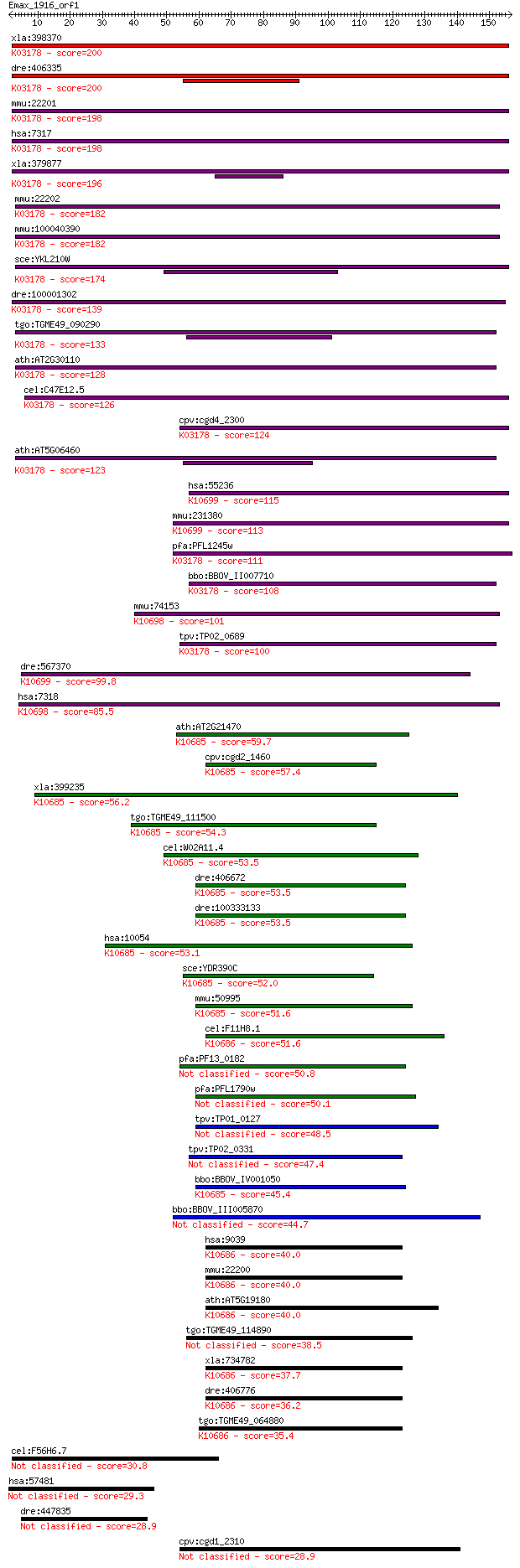
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1916_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
xla:398370 uba1-a, MGC68851, a1s9, a1s9t, a1st, amcx1, gxp1, p... 200 1e-51
dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:... 200 2e-51
mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-l... 198 5e-51
hsa:7317 UBA1, A1S9, A1S9T, A1ST, AMCX1, GXP1, MGC4781, POC20,... 198 6e-51
xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1, p... 196 2e-50
mmu:22202 Ube1y1, A1s9Y-1, Sby, Ube-2, Ube1ay, Ube1y, Ube1y-1;... 182 3e-46
mmu:100040390 ubiquitin-like modifier-activating enzyme 1 Y-li... 182 3e-46
sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme E1... 174 9e-44
dre:100001302 ubiquitin-like modifier-activating enzyme 1-like... 139 4e-33
tgo:TGME49_090290 ubiquitin-activating enzyme E1, putative ; K... 133 2e-31
ath:AT2G30110 ATUBA1; ATUBA1; ubiquitin activating enzyme/ ubi... 128 7e-30
cel:C47E12.5 uba-1; UBA (human ubiquitin) related family membe... 126 2e-29
cpv:cgd4_2300 ubiquitin-activating enzyme E1 (UBA) ; K03178 ub... 124 1e-28
ath:AT5G06460 ATUBA2; ATUBA2; ubiquitin activating enzyme/ ubi... 123 3e-28
hsa:55236 UBA6, E1-L2, FLJ10808, FLJ23367, MOP-4, UBE1L2; ubiq... 115 6e-26
mmu:231380 Uba6, 4930542H01, 5730469D23Rik, AU021846, AW124799... 113 3e-25
pfa:PFL1245w ubiquitin-activating enzyme E1, putative; K03178 ... 111 8e-25
bbo:BBOV_II007710 18.m06639; ubiquitin-activating enzyme E1; K... 108 5e-24
mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier ... 101 1e-21
tpv:TP02_0689 ubiquitin-protein ligase; K03178 ubiquitin-activ... 100 2e-21
dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquit... 99.8 3e-21
hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-lik... 85.5 6e-17
ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO acti... 59.7 3e-09
cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiq... 57.4 2e-08
xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-... 56.2 4e-08
tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.... 54.3 1e-07
cel:W02A11.4 uba-2; UBA (human ubiquitin) related family membe... 53.5 2e-07
dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354; u... 53.5 3e-07
dre:100333133 ubiquitin-like modifier activating enzyme 2-like... 53.5 3e-07
hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier a... 53.1 3e-07
sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterod... 52.0 8e-07
mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubi... 51.6 9e-07
cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family m... 51.6 1e-06
pfa:PF13_0182 ubiquitin-activating enzyme, putative 50.8 2e-06
pfa:PFL1790w ubiquitin-activating enzyme, putative 50.1 3e-06
tpv:TP01_0127 ubiquitin-protein ligase 48.5 9e-06
tpv:TP02_0331 ubiquitin activating enzyme, putatuve 47.4 2e-05
bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K106... 45.4 7e-05
bbo:BBOV_III005870 17.m07520; ThiF family protein 44.7 1e-04
hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin... 40.0 0.003
mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539, U... 40.0 0.003
ath:AT5G19180 ECR1; ECR1 (E1 C-terminal related 1); NEDD8 acti... 40.0 0.003
tgo:TGME49_114890 ubiquitin-activating enzyme E1, putative 38.5 0.010
xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier act... 37.7 0.017
dre:406776 uba3, ube1c, wu:fb75e04, wu:fc37b11, zgc:55528; ubi... 36.2 0.041
tgo:TGME49_064880 ubiquitin-activating enzyme, putative ; K106... 35.4 0.085
cel:F56H6.7 hypothetical protein 30.8 1.9
hsa:57481 KIAA1210 29.3 5.9
dre:447835 cx47.1, zgc:92348; connexin 47.1 28.9
cpv:cgd1_2310 hypothetical protein 28.9 8.0
> xla:398370 uba1-a, MGC68851, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1, uba1a, uba1b, ube, ube1, ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1060
Score = 200 bits (509), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 100/154 (64%), Positives = 122/154 (79%), Gaps = 4/154 (2%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
++P FTPK +KI V+D E+ +N++ D +T + L+ LP+ L G ++ PI+FEK
Sbjct 797 KVPEFTPKSGVKIHVSDQEI--QNAHASLD-DTRLEELKHALPTPESLGGFRMFPIDFEK 853
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDDTNFH+DFIVAASNLRAENY+IPPADRHKSKLIAG+IIPAIATTTA V GLV +ELYK
Sbjct 854 DDDTNFHMDFIVAASNLRAENYDIPPADRHKSKLIAGKIIPAIATTTAAVVGLVCLELYK 913
Query 122 LVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
++QG LE YKNGF+NLALPFFGFS PIA PK
Sbjct 914 IIQGH-RKLELYKNGFLNLALPFFGFSEPIAAPK 946
> dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:fj14g11,
zgc:66143; ubiquitin-like modifier activating enzyme
1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 200 bits (509), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 101/154 (65%), Positives = 120/154 (77%), Gaps = 4/154 (2%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
++P FTPK +KI V+D EL S N++V ++ + L+ LPS+ + K+ PIEFEK
Sbjct 795 KVPEFTPKSGVKIHVSDQELQSANASVD---DSRLEELKTLLPSLEASSQFKLCPIEFEK 851
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDDTNFH+DFIVAASNLRAENY+IPPADRHKSKLIAG+IIPAIATTTA V GLV +EL K
Sbjct 852 DDDTNFHMDFIVAASNLRAENYDIPPADRHKSKLIAGKIIPAIATTTAAVVGLVCLELLK 911
Query 122 LVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+VQG LE YKNGF+NLALPFF FS PIA PK
Sbjct 912 IVQGH-KKLESYKNGFMNLALPFFAFSEPIAAPK 944
Score = 33.5 bits (75), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 26/37 (70%), Gaps = 3/37 (8%)
Query 55 NPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPA-DR 90
+P+EF ++D H+D+I+AA+NL A +Y +P DR
Sbjct 750 HPLEFSTNND--LHMDYILAAANLYALSYGLPSCNDR 784
> mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 198 bits (504), Expect = 5e-51, Method: Composition-based stats.
Identities = 99/154 (64%), Positives = 122/154 (79%), Gaps = 4/154 (2%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
++P FTPK +KI V+D EL S N++V ++ + L+ LPS +L G K+ PI+FEK
Sbjct 795 QVPEFTPKSGVKIHVSDQELQSANASVD---DSRLEELKATLPSPDKLPGFKMYPIDFEK 851
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDD+NFH+DFIVAASNLRAENY+I PADRHKSKLIAG+IIPAIATTTA V GLV +ELYK
Sbjct 852 DDDSNFHMDFIVAASNLRAENYDISPADRHKSKLIAGKIIPAIATTTAAVVGLVCLELYK 911
Query 122 LVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+VQG L+ YKNGF+NLALPFFGFS P+A P+
Sbjct 912 VVQGH-QQLDSYKNGFLNLALPFFGFSEPLAAPR 944
> hsa:7317 UBA1, A1S9, A1S9T, A1ST, AMCX1, GXP1, MGC4781, POC20,
SMAX2, UBA1A, UBE1, UBE1X; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1058
Score = 198 bits (504), Expect = 6e-51, Method: Composition-based stats.
Identities = 99/154 (64%), Positives = 122/154 (79%), Gaps = 4/154 (2%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
++P FTPK +KI V+D EL S N++V ++ + L+ LPS +L G K+ PI+FEK
Sbjct 795 QVPEFTPKSGVKIHVSDQELQSANASVD---DSRLEELKATLPSPDKLPGFKMYPIDFEK 851
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDD+NFH+DFIVAASNLRAENY+IP ADRHKSKLIAG+IIPAIATTTA V GLV +ELYK
Sbjct 852 DDDSNFHMDFIVAASNLRAENYDIPSADRHKSKLIAGKIIPAIATTTAAVVGLVCLELYK 911
Query 122 LVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+VQG L+ YKNGF+NLALPFFGFS P+A P+
Sbjct 912 VVQGH-RQLDSYKNGFLNLALPFFGFSEPLAAPR 944
> xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1a, ube1, ube1x; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1059
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 98/154 (63%), Positives = 120/154 (77%), Gaps = 4/154 (2%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
++P FTP+ +KI V+D E+ +N++ D N + L+ LP+ L ++ PI+FEK
Sbjct 796 KVPEFTPRSGVKIHVSDQEI--QNAHASLDDN-RLEELKHTLPTPESLGSFRMFPIDFEK 852
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDDTNFH+DFIVAASNLRAENY+IPPADRHKSKLIAG+IIPAIATTTA V GLV +ELYK
Sbjct 853 DDDTNFHMDFIVAASNLRAENYDIPPADRHKSKLIAGKIIPAIATTTAAVVGLVCLELYK 912
Query 122 LVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
++QG LE YKNGF+NLALPFFGFS PIA PK
Sbjct 913 IIQGH-RKLESYKNGFLNLALPFFGFSEPIAAPK 945
Score = 28.9 bits (63), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 11/21 (52%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 65 TNFHIDFIVAASNLRAENYEI 85
T H+D+I+AA+NL A +Y I
Sbjct 759 TGLHVDYIMAAANLLASSYGI 779
> mmu:22202 Ube1y1, A1s9Y-1, Sby, Ube-2, Ube1ay, Ube1y, Ube1y-1;
ubiquitin-activating enzyme E1, Chr Y 1; K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 182 bits (462), Expect = 3e-46, Method: Composition-based stats.
Identities = 90/150 (60%), Positives = 113/150 (75%), Gaps = 3/150 (2%)
Query 3 IPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKD 62
+P F PK ++I V++ EL S ++ D +H + L+ LP+ +L G K+ PI+FEKD
Sbjct 795 VPKFAPKSGIRIHVSEQELQSTSATTIDD--SHLEELKTALPTPDKLLGFKMYPIDFEKD 852
Query 63 DDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKL 122
DD+NFH+DFIVAASNLRAENY I PADRHKSKLIAG+IIPAIATTT+ + GLV +ELYK+
Sbjct 853 DDSNFHMDFIVAASNLRAENYGISPADRHKSKLIAGKIIPAIATTTSAIVGLVCLELYKV 912
Query 123 VQGFTNTLEPYKNGFVNLALPFFGFSTPIA 152
VQG LE YKN F+NLALP F FS P+A
Sbjct 913 VQGH-QQLESYKNSFINLALPLFSFSAPLA 941
> mmu:100040390 ubiquitin-like modifier-activating enzyme 1 Y-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 182 bits (462), Expect = 3e-46, Method: Composition-based stats.
Identities = 90/150 (60%), Positives = 113/150 (75%), Gaps = 3/150 (2%)
Query 3 IPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKD 62
+P F PK ++I V++ EL S ++ D +H + L+ LP+ +L G K+ PI+FEKD
Sbjct 795 VPKFAPKSGIRIHVSEQELQSTSATTIDD--SHLEELKTALPTPDKLLGFKMYPIDFEKD 852
Query 63 DDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKL 122
DD+NFH+DFIVAASNLRAENY I PADRHKSKLIAG+IIPAIATTT+ + GLV +ELYK+
Sbjct 853 DDSNFHMDFIVAASNLRAENYGISPADRHKSKLIAGKIIPAIATTTSAIVGLVCLELYKV 912
Query 123 VQGFTNTLEPYKNGFVNLALPFFGFSTPIA 152
VQG LE YKN F+NLALP F FS P+A
Sbjct 913 VQGH-QQLESYKNSFINLALPLFSFSAPLA 941
> sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1024
Score = 174 bits (441), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 94/153 (61%), Positives = 110/153 (71%), Gaps = 3/153 (1%)
Query 3 IPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKD 62
IP FTP NLKI VND + + N+N + L LP S LAG K+ P++FEKD
Sbjct 767 IPEFTPNANLKIQVNDDDPDP-NANAANG-SDEIDQLVSSLPDPSTLAGFKLEPVDFEKD 824
Query 63 DDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKL 122
DDTN HI+FI A SN RA+NY I ADR K+K IAGRIIPAIATTT+LV+GLV +ELYKL
Sbjct 825 DDTNHHIEFITACSNCRAQNYFIETADRQKTKFIAGRIIPAIATTTSLVTGLVNLELYKL 884
Query 123 VQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+ T+ +E YKNGFVNLALPFFGFS PIA PK
Sbjct 885 IDNKTD-IEQYKNGFVNLALPFFGFSEPIASPK 916
Score = 33.1 bits (74), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 24/62 (38%), Positives = 30/62 (48%), Gaps = 9/62 (14%)
Query 49 LAGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEI--------PPADRHKSKLIAGRI 100
+G K P E D N H F+VA ++LRA NY I P D +KS +I I
Sbjct 708 WSGAKRAPTPLEFDIYNNDHFHFVVAGASLRAYNYGIKSDDSNSKPNVDEYKS-VIDHMI 766
Query 101 IP 102
IP
Sbjct 767 IP 768
> dre:100001302 ubiquitin-like modifier-activating enzyme 1-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1016
Score = 139 bits (350), Expect = 4e-33, Method: Composition-based stats.
Identities = 84/154 (54%), Positives = 111/154 (72%), Gaps = 2/154 (1%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVS-ELAGVKINPIEFE 60
++P FTPK ++KIAV D +LN EN + L++QL + +++P +FE
Sbjct 753 KVPEFTPKSSVKIAVTDQQLNEENEERKEEDKVKLGMLKEQLSKLQLRDRSFRMHPQDFE 812
Query 61 KDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELY 120
KDDD+NFH+D+IVAASNLRAENY+IP ADRHKSKLIAGRIIPAIATTTA ++GL+ +ELY
Sbjct 813 KDDDSNFHMDYIVAASNLRAENYDIPTADRHKSKLIAGRIIPAIATTTAAIAGLMCLELY 872
Query 121 KLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVP 154
KLVQG + + Y+N ++NLA +F FS P P
Sbjct 873 KLVQGHSK-ITSYRNAYINLATQYFVFSQPCPAP 905
> tgo:TGME49_090290 ubiquitin-activating enzyme E1, putative ;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1091
Score = 133 bits (335), Expect = 2e-31, Method: Composition-based stats.
Identities = 78/165 (47%), Positives = 101/165 (61%), Gaps = 19/165 (11%)
Query 3 IPVFTPKQNLKIAVNDSELNSENSNVPTDLNTH----------------FKNLQKQLPSV 46
IP FTPK+ L I +D+E N + P + L+K L +
Sbjct 807 IPQFTPKR-LHINTDDAE--KPNGSGPPGASFAAPHPSLSLSAEAEEEVVAGLEKHLLAT 863
Query 47 SELAGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIAT 106
++L + P+EFEKDDDTNFHID + AAS LRA NY+IP DR+K+K+IAGRIIPAIAT
Sbjct 864 ADLEKMVFVPVEFEKDDDTNFHIDLVHAASTLRAMNYKIPCCDRNKTKIIAGRIIPAIAT 923
Query 107 TTALVSGLVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
TTA+++GLV +EL K V LE +KN F NLALP + FS P+
Sbjct 924 TTAMITGLVSLELLKTVTYKQRKLEDFKNAFANLALPLWLFSEPM 968
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 29/46 (63%), Gaps = 3/46 (6%)
Query 56 PIEFEKDDDTNFHIDFIVAASNLRAENYEIPPA-DRHKSKLIAGRI 100
PI F+ +D + +DF+VAASNL A N+ +P D K + IA R+
Sbjct 762 PISFDANDPAS--LDFVVAASNLFAFNFGLPAVRDVSKIQAIAARV 805
> ath:AT2G30110 ATUBA1; ATUBA1; ubiquitin activating enzyme/ ubiquitin-protein
ligase; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1080
Score = 128 bits (322), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 65/154 (42%), Positives = 96/154 (62%), Gaps = 11/154 (7%)
Query 3 IPVFTPKQNLKIAVND-----SELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPI 57
+P F P+Q+ KI ++ + + +++ V DL + L ++ PI
Sbjct 824 VPDFEPRQDAKIVTDEKATTLTTASVDDAAVIDDLIAKIDQCRHNLS-----PDFRMKPI 878
Query 58 EFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFI 117
+FEKDDDTN+H+D I +N+RA NY IP D+ K+K IAGRIIPAIAT+TA+ +GLV +
Sbjct 879 QFEKDDDTNYHMDVIAGLANMRARNYSIPEVDKLKAKFIAGRIIPAIATSTAMATGLVCL 938
Query 118 ELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
ELYK++ G + +E Y+N F NLALP F + P+
Sbjct 939 ELYKVLDG-GHKVEAYRNTFANLALPLFSMAEPL 971
> cel:C47E12.5 uba-1; UBA (human ubiquitin) related family member
(uba-1); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1113
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 80/157 (50%), Positives = 111/157 (70%), Gaps = 7/157 (4%)
Query 6 FTPKQNLKIAVNDSELNSEN----SNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
F PK +KIAV D+E +N S++ D + + L+ +L +++ + K+N ++FEK
Sbjct 849 FEPKSGVKIAVTDAEAKEQNERGASSMIVDDDAAIEALKLKLATLNVKSTSKLNCVDFEK 908
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
DDD+N H++FI AASNLRAENY+I PADR ++K IAG+IIPAIATTTA V+GLV IELYK
Sbjct 909 DDDSNHHMEFITAASNLRAENYDILPADRMRTKQIAGKIIPAIATTTAAVAGLVCIELYK 968
Query 122 LVQ--GFTNT-LEPYKNGFVNLALPFFGFSTPIAVPK 155
+V G T +E +KN F+NL++PFF + PI PK
Sbjct 969 VVDANGIPKTPMERFKNTFLNLSMPFFSSAEPIGAPK 1005
> cpv:cgd4_2300 ubiquitin-activating enzyme E1 (UBA) ; K03178
ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1067
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 57/102 (55%), Positives = 74/102 (72%), Gaps = 0/102 (0%)
Query 54 INPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
I PIEFEKDDD+NFHIDF+ + +NLRA NY I DRHK K+IAGRIIPA+ATTTA+++G
Sbjct 845 IQPIEFEKDDDSNFHIDFMNSCANLRARNYSIKECDRHKCKMIAGRIIPAMATTTAMITG 904
Query 114 LVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
LV E K+ +E +KN F+NL+LP + + P+ PK
Sbjct 905 LVSFEALKVSSLGEYKIELFKNSFINLSLPLYVITEPLPAPK 946
> ath:AT5G06460 ATUBA2; ATUBA2; ubiquitin activating enzyme/ ubiquitin-protein
ligase; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1077
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 61/154 (39%), Positives = 95/154 (61%), Gaps = 11/154 (7%)
Query 3 IPVFTPKQNLKIAVND-----SELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPI 57
+P F PK++ I ++ S + +++ V +LN + L ++ I
Sbjct 821 VPDFEPKKDATIVTDEKATTLSTASVDDAAVIDELNAKLVRCRMSLQP-----EFRMKAI 875
Query 58 EFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFI 117
+FEKDDDTN+H+D I +N+RA NY +P D+ K+K IAGRIIPAIAT+TA+ +G V +
Sbjct 876 QFEKDDDTNYHMDMIAGLANMRARNYSVPEVDKLKAKFIAGRIIPAIATSTAMATGFVCL 935
Query 118 ELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
E+YK++ G ++ +E Y+N F NLALP F + P+
Sbjct 936 EMYKVLDG-SHKVEDYRNTFANLALPLFSMAEPV 968
Score = 32.3 bits (72), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 55 NPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSK 94
P++F D + HI+F++AAS LRAE + IP + K++
Sbjct 772 RPLQFSSTDLS--HINFVMAASILRAETFGIPTPEWAKTR 809
> hsa:55236 UBA6, E1-L2, FLJ10808, FLJ23367, MOP-4, UBE1L2; ubiquitin-like
modifier activating enzyme 6; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1052
Score = 115 bits (288), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 60/99 (60%), Positives = 71/99 (71%), Gaps = 2/99 (2%)
Query 57 IEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVF 116
+ FEKDDD N HIDFI AASNLRA+ Y I PADR K+K IAG+IIPAIATTTA VSGLV
Sbjct 847 LSFEKDDDHNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATTTATVSGLVA 906
Query 117 IELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
+E+ K+ G+ E YKN F+NLA+P F+ V K
Sbjct 907 LEMIKVTGGY--PFEAYKNCFLNLAIPIVVFTETTEVRK 943
> mmu:231380 Uba6, 4930542H01, 5730469D23Rik, AU021846, AW124799,
E1-L2, Ube1l2; ubiquitin-like modifier activating enzyme
6; K10699 ubiquitin-activating enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1053
Score = 113 bits (282), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 58/104 (55%), Positives = 74/104 (71%), Gaps = 2/104 (1%)
Query 52 VKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALV 111
+++ + FEKDDD N HIDFI AASNLRA+ Y I PADR K+K IAG+IIPAIAT+TA V
Sbjct 842 LQMTVLSFEKDDDRNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATSTAAV 901
Query 112 SGLVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIAVPK 155
SGLV +E+ K+ G+ + YKN F+NLA+P F+ V K
Sbjct 902 SGLVALEMIKVAGGY--PFDAYKNCFLNLAIPIIVFTETSEVRK 943
> pfa:PFL1245w ubiquitin-activating enzyme E1, putative; K03178
ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1140
Score = 111 bits (278), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 58/123 (47%), Positives = 79/123 (64%), Gaps = 18/123 (14%)
Query 52 VKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALV 111
+KINPIEF+KD+ TN H++FI A SNLRA NY+I D+ K+K++AG+IIPA+ATTT+++
Sbjct 898 IKINPIEFDKDEQTNLHVNFIYAFSNLRAINYKINTCDKLKAKIVAGKIIPALATTTSII 957
Query 112 SGLVFIELYKLVQGFTN------------------TLEPYKNGFVNLALPFFGFSTPIAV 153
+GLV IEL K V + N L +KN F+N ALP F FS P+
Sbjct 958 TGLVGIELLKYVNYYDNIQAYVKLSDEQRKKEKHDVLSYFKNAFINSALPLFLFSEPMPP 1017
Query 154 PKM 156
+M
Sbjct 1018 LRM 1020
> bbo:BBOV_II007710 18.m06639; ubiquitin-activating enzyme E1;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1007
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 48/95 (50%), Positives = 69/95 (72%), Gaps = 0/95 (0%)
Query 57 IEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVF 116
+EFEKDDDTN+HI+FI A +NLR +NY+I DR K+K+I+G+IIPAIATTT++++GLV
Sbjct 786 VEFEKDDDTNYHIEFIWATANLRCQNYDIDQCDRMKAKMISGKIIPAIATTTSMIAGLVM 845
Query 117 IELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
+E K + +E ++N F LA P + S P+
Sbjct 846 LEFVKTICYQKLKIEHFRNSFCCLATPLWLQSEPM 880
> mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier
activating enzyme 7; K10698 ubiquitin-activating enzyme E1-like
[EC:6.3.2.19]
Length=977
Score = 101 bits (251), Expect = 1e-21, Method: Composition-based stats.
Identities = 53/118 (44%), Positives = 78/118 (66%), Gaps = 6/118 (5%)
Query 40 QKQLPSVSEL-----AGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSK 94
Q+QL + E G + P+ F KDDD+NFH+DF+VAA++LR +NY I P + + K
Sbjct 749 QEQLKELQETLDDWRKGPPLKPVLFVKDDDSNFHVDFVVAATDLRCQNYGILPVNHARIK 808
Query 95 LIAGRIIPAIATTTALVSGLVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPIA 152
I GRIIPAIAT+TA+V+GL+ +ELYK+V G + +++ +++LA F S P A
Sbjct 809 QIVGRIIPAIATSTAVVAGLLGLELYKVVSGL-RSHGTFRHSYLHLAENHFIRSAPSA 865
> tpv:TP02_0689 ubiquitin-protein ligase; K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=999
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 48/98 (48%), Positives = 67/98 (68%), Gaps = 0/98 (0%)
Query 54 INPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
++ +EFEKDD+TN+HI+FI +AS LR NY I D+ K+KLI+G+IIPAIATTTA++ G
Sbjct 777 LDAVEFEKDDETNYHIEFIWSASVLRCRNYAIKECDKMKAKLISGKIIPAIATTTAMIGG 836
Query 114 LVFIELYKLVQGFTNTLEPYKNGFVNLALPFFGFSTPI 151
LV IE K + + ++N F LA P + S P+
Sbjct 837 LVTIEFLKALCYRNLKITHFRNAFACLATPIWLQSEPL 874
> dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1060
Score = 99.8 bits (247), Expect = 3e-21, Method: Composition-based stats.
Identities = 61/139 (43%), Positives = 85/139 (61%), Gaps = 13/139 (9%)
Query 5 VFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKDDD 64
V P Q LKI V+ E S + +N++ ++ + ++P+ FEKDDD
Sbjct 809 VKKPDQ-LKITVSSEEEREAISQLQEAINSNLVTPER----------LCMSPLFFEKDDD 857
Query 65 TNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKLVQ 124
TN H+DF+ +AS LRA Y I ADR ++K IAG+IIPAIAT+TA V+GLV +EL K+
Sbjct 858 TNGHMDFVASASALRARMYAIEAADRLQTKRIAGKIIPAIATSTAAVAGLVSMELIKIAG 917
Query 125 GFTNTLEPYKNGFVNLALP 143
G+ E +KN F NLA+P
Sbjct 918 GY--GFELFKNCFFNLAIP 934
> hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-like
modifier activating enzyme 7; K10698 ubiquitin-activating
enzyme E1-like [EC:6.3.2.19]
Length=1012
Score = 85.5 bits (210), Expect = 6e-17, Method: Composition-based stats.
Identities = 63/149 (42%), Positives = 89/149 (59%), Gaps = 15/149 (10%)
Query 4 PVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKDD 63
P+F NL++A +E E K L K L S G + P+ FEKDD
Sbjct 767 PIFA--SNLELASASAEFGPEQQ----------KELNKALEVWS--VGPPLKPLMFEKDD 812
Query 64 DTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKLV 123
D+NFH+DF+VAA++LR +NY IPP +R +SK I G+IIPAIATTTA V+GL+ +ELYK+V
Sbjct 813 DSNFHVDFVVAAASLRCQNYGIPPVNRAQSKRIVGQIIPAIATTTAAVAGLLGLELYKVV 872
Query 124 QGFTNTLEPYKNGFVNLALPFFGFSTPIA 152
G +++ +++LA + P A
Sbjct 873 SG-PRPRSAFRHSYLHLAENYLIRYMPFA 900
> ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO activating
enzyme; K10685 ubiquitin-like 1-activating enzyme E1
B [EC:6.3.2.19]
Length=625
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 50/72 (69%), Gaps = 2/72 (2%)
Query 53 KINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVS 112
+I + F+KDD ++F+ AA+N+RAE++ IP ++K IAG I+ A+ATT A+++
Sbjct 330 EIGHLTFDKDD--QLAVEFVTAAANIRAESFGIPLHSLFEAKGIAGNIVHAVATTNAIIA 387
Query 113 GLVFIELYKLVQ 124
GL+ IE K+++
Sbjct 388 GLIVIEAIKVLK 399
> cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=637
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/53 (49%), Positives = 36/53 (67%), Gaps = 0/53 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGL 114
D D +DF+ AASNLR+ N+ IP R + IAG I+PA+A+T A+VSG+
Sbjct 345 DKDNKDAMDFVSAASNLRSYNFHIPLQSRWSCQSIAGSIVPAVASTNAIVSGV 397
> xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-b,
uble1b-B; ubiquitin-like modifier activating enzyme 2;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=641
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 66/131 (50%), Gaps = 3/131 (2%)
Query 9 KQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEKDDDTNFH 68
K+N N+S L ++ ++ + K + ++ E K + E D D
Sbjct 290 KENCSEIQNESSLLGLKDQKVLNVASYAQLFSKSVETLREQLREKGDGAELVWDKDDVPA 349
Query 69 IDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYKLVQGFTN 128
+DF+ AA+NLR + + + K +AG IIPAIATT A++SGL+ +E K++ G T
Sbjct 350 MDFVTAAANLRMHIFSMNMKSKFDVKSMAGNIIPAIATTNAVISGLIVLEGLKILSGNT- 408
Query 129 TLEPYKNGFVN 139
E + F+N
Sbjct 409 --EQCRTVFLN 417
> tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.2.1.70);
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=730
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/78 (42%), Positives = 46/78 (58%), Gaps = 4/78 (5%)
Query 39 LQKQLPSVSELAGV--KINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLI 96
L++Q + E AG + I F+KDDD +DF+ AA+NLR N+ I R + +
Sbjct 384 LERQKTTERENAGTGKREAGIPFDKDDD--LAMDFVAAAANLRMHNFHIALKSRWFIQAV 441
Query 97 AGRIIPAIATTTALVSGL 114
AG IIPAIA T A+V+ L
Sbjct 442 AGSIIPAIAATNAVVAAL 459
> cel:W02A11.4 uba-2; UBA (human ubiquitin) related family member
(uba-2); K10685 ubiquitin-like 1-activating enzyme E1 B
[EC:6.3.2.19]
Length=582
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 48/80 (60%), Gaps = 1/80 (1%)
Query 49 LAGVKINP-IEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATT 107
L ++ P ++ D D + F+ A +N+RA+ + IP + K +AG IIPAIA+T
Sbjct 321 LEQIRAEPDVKLAFDKDHAIIMSFVAACANIRAKIFGIPMKSQFDIKAMAGNIIPAIAST 380
Query 108 TALVSGLVFIELYKLVQGFT 127
A+V+G++ E ++++G T
Sbjct 381 NAIVAGIIVTEAVRVIEGST 400
> dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354;
ubiquitin-like modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=640
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 42/65 (64%), Gaps = 2/65 (3%)
Query 59 FEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIE 118
++KDD +DF+ AASNLR + + R K +AG IIPAIATT A+++GL+ +E
Sbjct 339 WDKDDPP--AMDFVTAASNLRMNVFSMNMKSRFDVKSMAGNIIPAIATTNAVIAGLIVLE 396
Query 119 LYKLV 123
K++
Sbjct 397 ALKIL 401
> dre:100333133 ubiquitin-like modifier activating enzyme 2-like;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=642
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 42/65 (64%), Gaps = 2/65 (3%)
Query 59 FEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIE 118
++KDD +DF+ AASNLR + + R K +AG IIPAIATT A+++GL+ +E
Sbjct 339 WDKDDPP--AMDFVTAASNLRMNVFSMNMKSRFDVKSMAGNIIPAIATTNAVIAGLIVLE 396
Query 119 LYKLV 123
K++
Sbjct 397 ALKIL 401
> hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier
activating enzyme 2; K10685 ubiquitin-like 1-activating enzyme
E1 B [EC:6.3.2.19]
Length=640
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 52/95 (54%), Gaps = 0/95 (0%)
Query 31 DLNTHFKNLQKQLPSVSELAGVKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADR 90
D+ ++ + K + ++ K + E D D +DF+ +A+NLR + + R
Sbjct 314 DVKSYARLFSKSIETLRVHLAEKGDGAELIWDKDDPSAMDFVTSAANLRMHIFSMNMKSR 373
Query 91 HKSKLIAGRIIPAIATTTALVSGLVFIELYKLVQG 125
K +AG IIPAIATT A+++GL+ +E K++ G
Sbjct 374 FDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSG 408
> sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterodimer
with Aos1p to activate Smt3p (SUMO) before its conjugation
to proteins (sumoylation), which may play a role in protein
targeting; essential for viability; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=636
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 40/60 (66%), Gaps = 4/60 (6%)
Query 55 NPIEFEKDD-DTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
N IEF+KDD DT ++F+ A+N+R+ + IP K IAG IIPAIATT A+V+G
Sbjct 326 NHIEFDKDDADT---LEFVATAANIRSHIFNIPMKSVFDIKQIAGNIIPAIATTNAIVAG 382
> mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubiquitin-like
modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=638
Score = 51.6 bits (122), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 43/67 (64%), Gaps = 2/67 (2%)
Query 59 FEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIE 118
++KDD +DF+ +A+NLR + + R K +AG IIPAIATT A+++GL+ +E
Sbjct 342 WDKDDPP--AMDFVTSAANLRMHIFSMNMKSRFDIKSMAGNIIPAIATTNAVIAGLIVLE 399
Query 119 LYKLVQG 125
K++ G
Sbjct 400 GLKILSG 406
> cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family
member (rfl-1); K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=430
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 41/74 (55%), Gaps = 0/74 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D H+++++ ++LRAE Y I DR + + RIIPA+A+T A+++ +E K
Sbjct 244 DADDPIHVEWVLERASLRAEKYNIRGVDRRLTSGVLKRIIPAVASTNAVIAASCALEALK 303
Query 122 LVQGFTNTLEPYKN 135
L ++ Y N
Sbjct 304 LATNIAKPIDNYLN 317
> pfa:PF13_0182 ubiquitin-activating enzyme, putative
Length=1838
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 39/70 (55%), Gaps = 5/70 (7%)
Query 54 INPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
I +E KDD I+FI + +N+R ENY + I+ IIP+I T +++S
Sbjct 1274 IYNLESNKDD-----INFIYSVTNIRCENYNFKKLNMFDFLKISNNIIPSIVTIVSMISA 1328
Query 114 LVFIELYKLV 123
L F E+YK+V
Sbjct 1329 LAFFEMYKIV 1338
> pfa:PFL1790w ubiquitin-activating enzyme, putative
Length=686
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 42/68 (61%), Gaps = 2/68 (2%)
Query 59 FEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIE 118
F+KDDD I+FI + SN+R N+ I + + IAG IIPAI++T A+V+ L +
Sbjct 355 FDKDDDE--CINFITSISNIRMLNFCISQKSKFDIQSIAGNIIPAISSTNAIVASLQAFQ 412
Query 119 LYKLVQGF 126
L +++ F
Sbjct 413 LIHVIEYF 420
> tpv:TP01_0127 ubiquitin-protein ligase
Length=543
Score = 48.5 bits (114), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 44/77 (57%), Gaps = 4/77 (5%)
Query 59 FEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIE 118
F K+D +DF+ +A+NLR N+ I P + IAG I+PAIA T A+V+ ++
Sbjct 330 FSKNDQV--CMDFVSSAANLRMINFGIKPLSTWDVQSIAGAIVPAIAATNAIVASFQVVQ 387
Query 119 LYKLVQGF--TNTLEPY 133
L L++ NTL+ +
Sbjct 388 LLHLLKFLKSNNTLDSH 404
> tpv:TP02_0331 ubiquitin activating enzyme, putatuve
Length=1126
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 37/66 (56%), Gaps = 4/66 (6%)
Query 57 IEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVF 116
IE E +D T +F+ SNLRA + IP A + A I+PA++T ++ S L
Sbjct 827 IEEECNDST----EFVYLVSNLRARKFNIPEASKTNLVRKAKNIVPAVSTCVSIASSLSL 882
Query 117 IELYKL 122
+ELYKL
Sbjct 883 MELYKL 888
> bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=630
Score = 45.4 bits (106), Expect = 7e-05, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 2/65 (3%)
Query 59 FEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIE 118
F+K+D +DF+ +A+NLR N+ IP + IAG I PAIA T A+V+ ++
Sbjct 407 FDKEDP--ICVDFVSSAANLRMINFNIPHLSTWDVQSIAGSITPAIAATNAIVAATQVMQ 464
Query 119 LYKLV 123
L L+
Sbjct 465 LIHLL 469
> bbo:BBOV_III005870 17.m07520; ThiF family protein
Length=1009
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 50/96 (52%), Gaps = 5/96 (5%)
Query 52 VKINPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALV 111
V++N + F D++ F+ AASN+RA + + D +A IIPAI+TT +
Sbjct 724 VELNALTF--DENMKDDCMFLYAASNIRAHKFGLNQKDMASVIKVAKGIIPAISTTVGVA 781
Query 112 SGLVFIELYK-LVQGFTNTLEPYKNGFVNLALPFFG 146
+ + +ELYK L NT+E K + P+FG
Sbjct 782 ASMAILELYKALYLMENNTIECEKRH--DDIDPYFG 815
> hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin-like
modifier activating enzyme 3; K10686 ubiquitin-activating
enzyme E1 C [EC:6.3.2.19]
Length=463
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 33/61 (54%), Gaps = 0/61 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D HI +I S RA Y I ++ + RIIPA+A+T A+++ + E++K
Sbjct 271 DGDDPEHIQWIFQKSLERASQYNIRGVTYRLTQGVVKRIIPAVASTNAVIAAVCATEVFK 330
Query 122 L 122
+
Sbjct 331 I 331
> mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539,
Ube1c; ubiquitin-like modifier activating enzyme 3; K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=448
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 33/61 (54%), Gaps = 0/61 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D HI +I S RA Y I ++ + RIIPA+A+T A+++ + E++K
Sbjct 257 DGDDPEHIQWIFQKSIERASQYNIRGVTYRLTQGVVKRIIPAVASTNAVIAAVCATEVFK 316
Query 122 L 122
+
Sbjct 317 I 317
> ath:AT5G19180 ECR1; ECR1 (E1 C-terminal related 1); NEDD8 activating
enzyme/ protein heterodimerization/ small protein activating
enzyme; K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=454
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 37/72 (51%), Gaps = 0/72 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D H+ ++ + RAE + IP ++ + IIPAIA+T A++S +E K
Sbjct 246 DPDEPEHMKWVYDEAIRRAELFGIPGVTYSLTQGVVKNIIPAIASTNAIISAACALETLK 305
Query 122 LVQGFTNTLEPY 133
+V + TL Y
Sbjct 306 IVSACSKTLVNY 317
> tgo:TGME49_114890 ubiquitin-activating enzyme E1, putative
Length=2759
Score = 38.5 bits (88), Expect = 0.010, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 44/73 (60%), Gaps = 7/73 (9%)
Query 56 PIEFEKDDDTNFHIDFIVAASNLRAENY---EIPPADRHKSKLIAGRIIPAIATTTALVS 112
P+ + KD + H+ F+ A + LRA + ++P D + ++GRI+PA +T T + +
Sbjct 2151 PMTYNKD--SPVHLSFLTATARLRARCFLFSDLP--DLLAVQQLSGRIVPATSTATTVAA 2206
Query 113 GLVFIELYKLVQG 125
GL +E+Y+LVQ
Sbjct 2207 GLAALEVYRLVQA 2219
> xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier activating
enzyme 3; K10686 ubiquitin-activating enzyme E1 C
[EC:6.3.2.19]
Length=461
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 33/61 (54%), Gaps = 0/61 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D HI++I S RA + I ++ + RIIPA+A+T A+++ E++K
Sbjct 270 DGDDPEHIEWIFTNSLERANQFNIRGVTYRLTQGVVKRIIPAVASTNAVIAAACATEVFK 329
Query 122 L 122
+
Sbjct 330 I 330
> dre:406776 uba3, ube1c, wu:fb75e04, wu:fc37b11, zgc:55528; ubiquitin-like
modifier activating enzyme 3 (EC:6.3.2.-); K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=462
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 62 DDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIELYK 121
D D HI ++ S RA + I ++ + RIIPA+A+T A+++ E++K
Sbjct 270 DGDDPKHIQWVYQKSLERAAEFNITGVTYRLTQGVVKRIIPAVASTNAVIAAACATEVFK 329
Query 122 L 122
+
Sbjct 330 I 330
> tgo:TGME49_064880 ubiquitin-activating enzyme, putative ; K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=668
Score = 35.4 bits (80), Expect = 0.085, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 60 EKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSGLVFIEL 119
E D D H+ ++ + RAE + I + + RIIPA+A+T A+++ ++ E
Sbjct 343 EFDGDNPEHLQWLYERAKQRAETFGIQGVTYRLTLGVTKRIIPAVASTNAIIAAMLVEEA 402
Query 120 YKL 122
K+
Sbjct 403 LKI 405
> cel:F56H6.7 hypothetical protein
Length=804
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 2 EIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPSVSELAGVKINPIEFEK 61
E + P++N I + +LN V T + NL Q PS ++LA + F+K
Sbjct 442 EFDLLKPRENQSIKRAEQKLNYFGDIVDWCNETGYSNLSNQFPSATQLAKQHGDTYVFQK 501
Query 62 DDDT 65
D ++
Sbjct 502 DQNS 505
> hsa:57481 KIAA1210
Length=1709
Score = 29.3 bits (64), Expect = 5.9, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 1 EEIPVFTPKQNLKIAVNDSELNSENSNVPTDLNTHFKNLQKQLPS 45
E++P+ P Q L N E++S ++N P + N + L + PS
Sbjct 871 EDLPLRHPAQALGKPKNQQEVSSASNNTPEEQNDFMQQLPSRCPS 915
> dre:447835 cx47.1, zgc:92348; connexin 47.1
Length=409
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query 5 VFTPKQNLKIAVNDSELNSENSNVPTDLNT---HFKNLQKQL 43
+ P QN+ + + S + N+PTDL T H + Q+QL
Sbjct 305 IVLPDQNMDREIAEQHCTSPDENIPTDLATLHHHLRVAQEQL 346
> cpv:cgd1_2310 hypothetical protein
Length=163
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 38/88 (43%), Gaps = 8/88 (9%)
Query 54 INPIEFEKDDDTNFHIDFIVAASNLRAENYEIPPADRHKSKLIAGRIIPAIATTTALVSG 113
I IE K D I+F + SN+R + Y +SKL + + +L+S
Sbjct 8 IMDIEKNKLDKIKQQIEFYFSDSNIRHDKY-------FRSKLTEYKYLHNFGIPISLIST 60
Query 114 L-VFIELYKLVQGFTNTLEPYKNGFVNL 140
IEL VQ N+L K FVN+
Sbjct 61 FNKMIELNASVQDIINSLSSSKVVFVNI 88
Lambda K H
0.317 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3451799900
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40