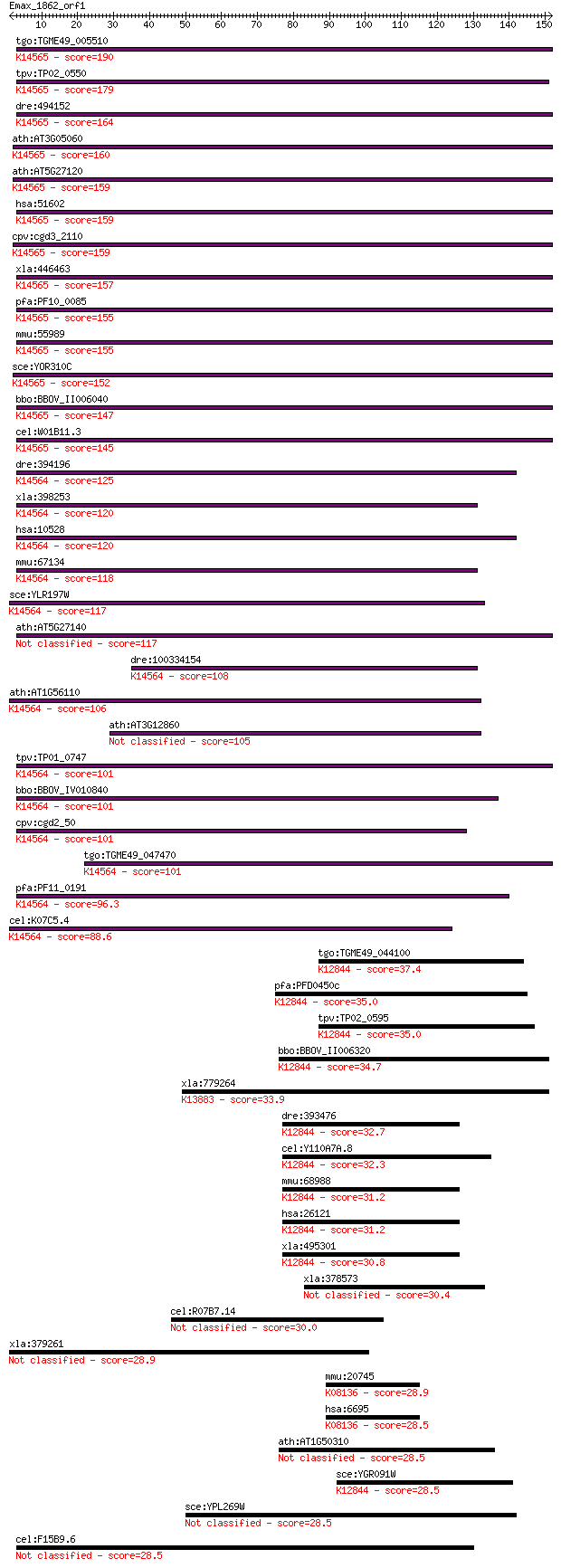
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1862_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005510 nucleolar protein NOP5, putative ; K14565 nu... 190 1e-48
tpv:TP02_0550 nuclear protein; K14565 nucleolar protein 58 179 3e-45
dre:494152 nop58, NOP5/NOP58, nol5, wu:fa93b01, zgc:65841; NOP... 164 1e-40
ath:AT3G05060 SAR DNA-binding protein, putative; K14565 nucleo... 160 2e-39
ath:AT5G27120 SAR DNA-binding protein, putative; K14565 nucleo... 159 2e-39
hsa:51602 NOP58, NOP5, NOP5/NOP58; NOP58 ribonucleoprotein hom... 159 2e-39
cpv:cgd3_2110 nucleolar protein NOP5/NOP58-like pre-mRNA splic... 159 4e-39
xla:446463 nop58-a, MGC78950, nop5, nop5/nop58, nop58; NOP58 r... 157 1e-38
pfa:PF10_0085 nucleolar protein NOP5, putative; K14565 nucleol... 155 5e-38
mmu:55989 Nop58, MGC105209, MSSP, Nol5, SIK, nop5; NOP58 ribon... 155 5e-38
sce:YOR310C NOP58, NOP5; Nop58p; K14565 nucleolar protein 58 152 5e-37
bbo:BBOV_II006040 18.m06502; nucleolar protein NOP5; K14565 nu... 147 9e-36
cel:W01B11.3 nol-5; NucleOLar protein family member (nol-5); K... 145 7e-35
dre:394196 nop56, nol5a; NOP56 ribonucleoprotein homolog; K145... 125 5e-29
xla:398253 nop56, MGC130628, nol5a, xnop56; NOP56 ribonucleopr... 120 1e-27
hsa:10528 NOP56, NOL5A; NOP56 ribonucleoprotein homolog (yeast... 120 2e-27
mmu:67134 Nop56, 2310044F10Rik, Nol5a; NOP56 ribonucleoprotein... 118 8e-27
sce:YLR197W NOP56, SIK1; Essential evolutionarily-conserved nu... 117 9e-27
ath:AT5G27140 SAR DNA-binding protein, putative 117 1e-26
dre:100334154 NOP56-like; K14564 nucleolar protein 56 108 5e-24
ath:AT1G56110 NOP56; NOP56 (Arabidopsis homolog of nucleolar p... 106 2e-23
ath:AT3G12860 nucleolar protein Nop56, putative 105 5e-23
tpv:TP01_0747 hypothetical protein; K14564 nucleolar protein 56 101 7e-22
bbo:BBOV_IV010840 23.m05998; nucleolar protein Nop56; K14564 n... 101 8e-22
cpv:cgd2_50 SIK1 nucleolar protein Nop56 ; K14564 nucleolar pr... 101 9e-22
tgo:TGME49_047470 nucleolar protein 5A, putative ; K14564 nucl... 101 1e-21
pfa:PF11_0191 Nop56/Sik1-like protein, putative; K14564 nucleo... 96.3 3e-20
cel:K07C5.4 hypothetical protein; K14564 nucleolar protein 56 88.6
tgo:TGME49_044100 putative snoRNA binding domain-containing pr... 37.4 0.016
pfa:PFD0450c pre-mRNA splicing factor, putative; K12844 U4/U6 ... 35.0 0.087
tpv:TP02_0595 hypothetical protein; K12844 U4/U6 small nuclear... 35.0 0.088
bbo:BBOV_II006320 18.m06519; pre-mRNA processing ribonucleopro... 34.7 0.11
xla:779264 rilp, MGC154468; Rab interacting lysosomal protein;... 33.9 0.21
dre:393476 prpf31, MGC66177, zgc:66177; PRP31 pre-mRNA process... 32.7 0.46
cel:Y110A7A.8 hypothetical protein; K12844 U4/U6 small nuclear... 32.3 0.59
mmu:68988 Prpf31, 1500019O16Rik, 2810404O06Rik, AW554706, PRP3... 31.2 1.3
hsa:26121 PRPF31, DKFZp566J153, NY-BR-99, PRP31, RP11; PRP31 p... 31.2 1.3
xla:495301 prpf31, prp31, rp11; PRP31 pre-mRNA processing fact... 30.8 1.6
xla:378573 slc13a2, NaDC-2, nact, nadc-1, nadc1, nadc2-a, sdct... 30.4 2.1
cel:R07B7.14 nhr-207; Nuclear Hormone Receptor family member (... 30.0 3.1
xla:379261 MGC53392; similar to angiopoietin-like factor 28.9 6.3
mmu:20745 Spock1, Spock, Ticn1, testican; sparc/osteonectin, c... 28.9 7.4
hsa:6695 SPOCK1, FLJ37170, SPOCK, TESTICAN, TIC1; sparc/osteon... 28.5 8.1
ath:AT1G50310 STP9; STP9 (SUGAR TRANSPORTER 9); carbohydrate t... 28.5 8.2
sce:YGR091W PRP31; Prp31p; K12844 U4/U6 small nuclear ribonucl... 28.5 8.6
sce:YPL269W KAR9; Kar9p 28.5 9.1
cel:F15B9.6 hypothetical protein 28.5 9.5
> tgo:TGME49_005510 nucleolar protein NOP5, putative ; K14565
nucleolar protein 58
Length=490
Score = 190 bits (483), Expect = 1e-48, Method: Composition-based stats.
Identities = 84/149 (56%), Positives = 116/149 (77%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L V+DK L I+ + I+V+F+P+ +E+IRGIK LS+L+ GL +DR +M+M L+H L
Sbjct 91 LAVSDKALGAAIRNKFGIDVVFTPTTHEIIRGIKEQLSNLLDGLSAKDRQQMAMSLAHSL 150
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
NR KL+F+P+KLDTMIIQA+ L+++LD+ELNNF MR+KEWYGWHFPEL+KI+ D+L+Y K
Sbjct 151 NRFKLKFSPEKLDTMIIQAVALIDDLDRELNNFAMRLKEWYGWHFPELSKIVTDNLIYAK 210
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
+QLI FRSN + L LLP EI E++
Sbjct 211 TVQLIGFRSNTRNVELSPLLPDEIAAEVK 239
> tpv:TP02_0550 nuclear protein; K14565 nucleolar protein 58
Length=425
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 78/148 (52%), Positives = 110/148 (74%), Gaps = 0/148 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L + DK L I+K LNINV F+ E+IRG+++ L++GL ++D M++ LSH L
Sbjct 90 LAICDKALGMEIQKNLNINVCFNQKTSEIIRGLRMQFHELVSGLSEEDTRSMALSLSHSL 149
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
R KL+F+PDK+D MI+QAIGLL++LD+E+N F MR+KEWYGWHFPEL+KI+ D+LLY +
Sbjct 150 TRFKLKFSPDKVDVMIVQAIGLLDDLDREVNKFGMRLKEWYGWHFPELDKIVSDNLLYAR 209
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEI 150
+I+ I R NAK A L +LP ++C+EI
Sbjct 210 VIKRIGMRENAKNANLSDILPEDVCKEI 237
> dre:494152 nop58, NOP5/NOP58, nol5, wu:fa93b01, zgc:65841; NOP58
ribonucleoprotein homolog (yeast); K14565 nucleolar protein
58
Length=529
Score = 164 bits (414), Expect = 1e-40, Method: Composition-based stats.
Identities = 75/149 (50%), Positives = 106/149 (71%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L + D L IK++LN++ + SP+ E++RGI+ + LITGL ++ MS+ L+H L
Sbjct 88 LAITDAKLGGVIKEKLNLSCVHSPAVAELMRGIRNQMEGLITGLPAREIAAMSLGLAHSL 147
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R KL+F+PDK+DTMI+QAI LL++LDKELNN+IMR +EWYGWHFPE+ KII D+L YCK
Sbjct 148 SRYKLKFSPDKVDTMIVQAISLLDDLDKELNNYIMRCREWYGWHFPEIGKIITDNLAYCK 207
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
++ I R+N L LP EI E++
Sbjct 208 TVRKIGDRTNVATTELSEFLPEEIEAEVK 236
> ath:AT3G05060 SAR DNA-binding protein, putative; K14565 nucleolar
protein 58
Length=533
Score = 160 bits (404), Expect = 2e-39, Method: Composition-based stats.
Identities = 76/150 (50%), Positives = 106/150 (70%), Gaps = 0/150 (0%)
Query 2 TLIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHG 61
TL VAD L N IK++L I+ + + + E++RG++ + LI+GL DQD MS+ LSH
Sbjct 85 TLAVADSKLGNVIKEKLKIDCIHNNAVMELLRGVRSQFTELISGLGDQDLAPMSLGLSHS 144
Query 62 LNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYC 121
L R KL+F+ DK+DTMIIQAIGLL++LDKELN + MR++EWYGWHFPEL KII D++LY
Sbjct 145 LARYKLKFSSDKVDTMIIQAIGLLDDLDKELNTYAMRVREWYGWHFPELAKIISDNILYA 204
Query 122 KIIQLIQFRSNAKYAPLETLLPHEICEEIR 151
K ++L+ R NA +L EI +++
Sbjct 205 KSVKLMGNRVNAAKLDFSEILADEIEADLK 234
> ath:AT5G27120 SAR DNA-binding protein, putative; K14565 nucleolar
protein 58
Length=533
Score = 159 bits (403), Expect = 2e-39, Method: Composition-based stats.
Identities = 77/150 (51%), Positives = 106/150 (70%), Gaps = 0/150 (0%)
Query 2 TLIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHG 61
TL VAD L N IK++L I + + + E++RGI+ L+ LI+GL DQD MS+ LSH
Sbjct 84 TLAVADSKLGNIIKEKLKIVCVHNNAVMELLRGIRSQLTELISGLGDQDLGPMSLGLSHS 143
Query 62 LNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYC 121
L R KL+F+ DK+DTMIIQAIGLL++LDKELN + MR++EW+GWHFPEL KI+ D++LY
Sbjct 144 LARYKLKFSSDKVDTMIIQAIGLLDDLDKELNTYAMRVREWFGWHFPELAKIVQDNILYA 203
Query 122 KIIQLIQFRSNAKYAPLETLLPHEICEEIR 151
K ++L+ R NA +L EI E++
Sbjct 204 KAVKLMGNRINAAKLDFSEILADEIEAELK 233
> hsa:51602 NOP58, NOP5, NOP5/NOP58; NOP58 ribonucleoprotein homolog
(yeast); K14565 nucleolar protein 58
Length=529
Score = 159 bits (403), Expect = 2e-39, Method: Composition-based stats.
Identities = 74/149 (49%), Positives = 105/149 (70%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L VAD L IK++LN++ + SP E++RGI+ + LI G++ ++ M + L+H L
Sbjct 87 LAVADAKLGGVIKEKLNLSCIHSPVVNELMRGIRSQMDGLIPGVEPREMAAMCLGLAHSL 146
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R +L+F+ DK+DTMI+QAI LL++LDKELNN+IMR +EWYGWHFPEL KII D+L YCK
Sbjct 147 SRYRLKFSADKVDTMIVQAISLLDDLDKELNNYIMRCREWYGWHFPELGKIISDNLTYCK 206
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
+Q + R N A L LLP E+ E++
Sbjct 207 CLQKVGDRKNYASAKLSELLPEEVEAEVK 235
> cpv:cgd3_2110 nucleolar protein NOP5/NOP58-like pre-mRNA splicinig
factor prp31 ; K14565 nucleolar protein 58
Length=467
Score = 159 bits (401), Expect = 4e-39, Method: Composition-based stats.
Identities = 76/153 (49%), Positives = 110/153 (71%), Gaps = 3/153 (1%)
Query 2 TLIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHG 61
+L VADK L IK++ I+V+++P EV+RGI+ L+ L+TGL ++D M++ LSH
Sbjct 89 SLAVADKVLGGLIKQKYEIDVVYNPKMQEVMRGIRNQLTDLLTGLTEKDMKTMALSLSHS 148
Query 62 LNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYC 121
L R KL+F+P+K+DTMIIQA+ LL++LD+ELNN+ MR+KEWYGWHFPEL KII D +Y
Sbjct 149 LGRFKLKFSPEKIDTMIIQAVALLDDLDRELNNYAMRLKEWYGWHFPELGKIISDRDVYA 208
Query 122 KIIQLIQFRSNAKYAPLETL---LPHEICEEIR 151
I++I FR + A L++ +P E+ EI+
Sbjct 209 NCIKVIGFRHCTRDANLQSPPCNIPSEMEAEIK 241
> xla:446463 nop58-a, MGC78950, nop5, nop5/nop58, nop58; NOP58
ribonucleoprotein homolog; K14565 nucleolar protein 58
Length=534
Score = 157 bits (397), Expect = 1e-38, Method: Composition-based stats.
Identities = 72/149 (48%), Positives = 104/149 (69%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L +AD L IK ++NI+ + + E++RGI+ + LITG+ ++ MS+ L+H L
Sbjct 88 LAIADAKLGGVIKDKVNISCVHTSMVTELMRGIRNQVDGLITGISSREMAAMSLGLAHSL 147
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R KL+F+PDK+DTMI+QAI LL++LDKELNN+IMR +EWYGWHFPEL K+I D+L YCK
Sbjct 148 SRYKLKFSPDKVDTMIVQAISLLDDLDKELNNYIMRCREWYGWHFPELGKVITDNLAYCK 207
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
++ + R N L LLP E+ E++
Sbjct 208 CVRAVGDRINFATFDLSELLPEEVETEVK 236
> pfa:PF10_0085 nucleolar protein NOP5, putative; K14565 nucleolar
protein 58
Length=469
Score = 155 bits (392), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 78/149 (52%), Positives = 107/149 (71%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L + DK L IKK+ NINV ++ S E+IRGI+ HL L+ G++++D + + LSH L
Sbjct 90 LALCDKTLGGLIKKKYNINVTYTNSTQEIIRGIRTHLYELLVGVNEEDSKLLKLSLSHSL 149
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
NR KL+F+ DK+D MI+QA+GLLE+LDKE+N F MR+KEWYGWHFPEL K+I D+ +Y K
Sbjct 150 NRFKLKFSADKVDVMIVQAVGLLEDLDKEINVFSMRLKEWYGWHFPELGKVITDNKIYAK 209
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
++LI FR+NAK L EI +EI+
Sbjct 210 CVKLIGFRNNAKNVNLLEETTEEIQKEIK 238
> mmu:55989 Nop58, MGC105209, MSSP, Nol5, SIK, nop5; NOP58 ribonucleoprotein
homolog (yeast); K14565 nucleolar protein 58
Length=536
Score = 155 bits (391), Expect = 5e-38, Method: Composition-based stats.
Identities = 72/149 (48%), Positives = 103/149 (69%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L VAD L IK++LN++ + SP E++RGI+ + LI G++ ++ M + L+H L
Sbjct 87 LAVADAKLGGVIKEKLNLSCIHSPVVNELMRGIRSQMDGLIPGVEPREMAAMCLGLAHSL 146
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R +L+F+ DK+DTMI+QAI LL++LDKELNN+IMR +EWYGWHFPEL KII D+L YCK
Sbjct 147 SRYRLKFSADKVDTMIVQAISLLDDLDKELNNYIMRCREWYGWHFPELGKIISDNLTYCK 206
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
+Q + R N A L L E+ E++
Sbjct 207 CLQKVGDRKNYASASLSEFLSEEVEAEVK 235
> sce:YOR310C NOP58, NOP5; Nop58p; K14565 nucleolar protein 58
Length=511
Score = 152 bits (383), Expect = 5e-37, Method: Composition-based stats.
Identities = 75/151 (49%), Positives = 100/151 (66%), Gaps = 1/151 (0%)
Query 2 TLIVADKNLANNIKK-QLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSH 60
TLIV++ LAN I K LN NV+ ++ R IK +L L+ G+ D D +MS+ L+H
Sbjct 86 TLIVSETKLANAINKLGLNFNVVSDAVTLDIYRAIKEYLPELLPGMSDNDLSKMSLGLAH 145
Query 61 GLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLY 120
+ R KL+F+ DK+D MIIQAI LL++LDKELN + MR KEWYGWHFPEL KI+ DS+ Y
Sbjct 146 SIGRHKLKFSADKVDVMIIQAIALLDDLDKELNTYAMRCKEWYGWHFPELAKIVTDSVAY 205
Query 121 CKIIQLIQFRSNAKYAPLETLLPHEICEEIR 151
+II + RS A L +LP EI E ++
Sbjct 206 ARIILTMGIRSKASETDLSEILPEEIEERVK 236
> bbo:BBOV_II006040 18.m06502; nucleolar protein NOP5; K14565
nucleolar protein 58
Length=439
Score = 147 bits (372), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 71/149 (47%), Positives = 106/149 (71%), Gaps = 0/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L V DK+LA I+K+L I+V+++P+ E+ RG+K L++G+ + D MS+ LSH L
Sbjct 90 LGVCDKSLAIEIQKKLKIDVVYNPNTLEIARGLKGQFFELVSGISESDARSMSLSLSHSL 149
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
R KL+F+PDK+D M++QAIGLL++LD+E NNF MR+KEWYGWHFPEL+ I+ D LY +
Sbjct 150 ARFKLKFSPDKVDIMVVQAIGLLDDLDREANNFGMRLKEWYGWHFPELSHIVPDMTLYSR 209
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
++ I R + LE+ LP ++ +EI+
Sbjct 210 AVRQIGIRGSTSLDELESFLPKDVVDEIK 238
> cel:W01B11.3 nol-5; NucleOLar protein family member (nol-5);
K14565 nucleolar protein 58
Length=487
Score = 145 bits (365), Expect = 7e-35, Method: Composition-based stats.
Identities = 65/149 (43%), Positives = 102/149 (68%), Gaps = 1/149 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L V D L N IK++L++N + S E++RG++ H+ L+ ++ + M++ ++H L
Sbjct 86 LAVGDAKLGNLIKEKLSLNCVHDSSINELMRGVRAHIEDLLAE-HKEEMNAMNLAVAHSL 144
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
R K++F P+K+DTMI+QA+ LL++LDKELNN++MR++EWYGWHFPEL K I D Y K
Sbjct 145 ARYKVKFNPEKIDTMIVQAVSLLDDLDKELNNYVMRVREWYGWHFPELGKTIQDHQAYAK 204
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
II+ I R N L ++LP E+ E+++
Sbjct 205 IIKAIGMRQNCINTDLSSILPEELEEKVK 233
> dre:394196 nop56, nol5a; NOP56 ribonucleoprotein homolog; K14564
nucleolar protein 56
Length=494
Score = 125 bits (314), Expect = 5e-29, Method: Composition-based stats.
Identities = 57/139 (41%), Positives = 89/139 (64%), Gaps = 0/139 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L V+D L ++++LN+++ E+IRG++LH SL+ GL Q + + L H
Sbjct 94 LGVSDAKLGAALQEELNLSIQTGGVVAEIIRGVRLHFHSLVKGLTAQAASKAQLGLGHSY 153
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R K++F ++ D MIIQ+I LL++LDK++N F MR++EWYG+HFPEL KI+ D+ YCK
Sbjct 154 SRAKVKFNVNRADNMIIQSIALLDQLDKDINTFSMRVREWYGYHFPELIKIVSDNSTYCK 213
Query 123 IIQLIQFRSNAKYAPLETL 141
+ +LI R LE +
Sbjct 214 LAKLIGNRKELSEEMLEAM 232
> xla:398253 nop56, MGC130628, nol5a, xnop56; NOP56 ribonucleoprotein
homolog; K14564 nucleolar protein 56
Length=532
Score = 120 bits (302), Expect = 1e-27, Method: Composition-based stats.
Identities = 54/128 (42%), Positives = 84/128 (65%), Gaps = 0/128 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L VAD + I+++L I + E++RGI+LH SL+ GL Q + + L H
Sbjct 93 LGVADAKIGAAIQEELKIPCQTGGAVVEILRGIRLHFHSLVKGLTAQSASKAQLGLGHSY 152
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R K++F +++D MIIQ+I LL++LDK++N F MR++EWYG+HFPEL KI+ D+ YC+
Sbjct 153 SRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRVREWYGYHFPELIKIVSDNYTYCR 212
Query 123 IIQLIQFR 130
+ + I R
Sbjct 213 MAKFIGNR 220
> hsa:10528 NOP56, NOL5A; NOP56 ribonucleoprotein homolog (yeast);
K14564 nucleolar protein 56
Length=594
Score = 120 bits (301), Expect = 2e-27, Method: Composition-based stats.
Identities = 56/139 (40%), Positives = 86/139 (61%), Gaps = 0/139 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L V D + I+++L N E++RG++LH +L+ GL D + + L H
Sbjct 93 LGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHFHNLVKGLTDLSACKAQLGLGHSY 152
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R K++F +++D MIIQ+I LL++LDK++N F MR++EWYG+HFPEL KII+D+ YC+
Sbjct 153 SRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRVREWYGYHFPELVKIINDNATYCR 212
Query 123 IIQLIQFRSNAKYAPLETL 141
+ Q I R LE L
Sbjct 213 LAQFIGNRRELNEDKLEKL 231
> mmu:67134 Nop56, 2310044F10Rik, Nol5a; NOP56 ribonucleoprotein
homolog (yeast); K14564 nucleolar protein 56
Length=580
Score = 118 bits (295), Expect = 8e-27, Method: Composition-based stats.
Identities = 52/128 (40%), Positives = 83/128 (64%), Gaps = 0/128 (0%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L V D + I+++L N E++RG++LH +L+ GL D + + L H
Sbjct 93 LGVGDPKIGAAIQEELGYNCQTGGVIAEILRGVRLHFHNLVKGLTDLSACKAQLGLGHSY 152
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
+R K++F +++D MIIQ+I LL++LDK++N F MR++EWYG+HFPEL KI++D+ YC+
Sbjct 153 SRAKVKFNVNRVDNMIIQSISLLDQLDKDINTFSMRVREWYGYHFPELVKIVNDNATYCR 212
Query 123 IIQLIQFR 130
+ Q I R
Sbjct 213 LAQFIGNR 220
> sce:YLR197W NOP56, SIK1; Essential evolutionarily-conserved
nucleolar protein component of the box C/D snoRNP complexes
that direct 2'-O-methylation of pre-rRNA during its maturation;
overexpression causes spindle orientation defects; K14564
nucleolar protein 56
Length=504
Score = 117 bits (294), Expect = 9e-27, Method: Composition-based stats.
Identities = 54/133 (40%), Positives = 88/133 (66%), Gaps = 1/133 (0%)
Query 1 LTLIVADKNLANNIKKQLN-INVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLS 59
+TL ++DKNL +IK++ ++ + + ++IRG++LH L GL D R + L
Sbjct 96 ITLAISDKNLGPSIKEEFPYVDCISNELAQDLIRGVRLHGEKLFKGLQSGDLERAQLGLG 155
Query 60 HGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLL 119
H +R K++F+ K D IIQAI LL++LDK++N F MR+KEWYGWHFPEL K++ D+
Sbjct 156 HAYSRAKVKFSVQKNDNHIIQAIALLDQLDKDINTFAMRVKEWYGWHFPELAKLVPDNYT 215
Query 120 YCKIIQLIQFRSN 132
+ K++ I+ +++
Sbjct 216 FAKLVLFIKDKAS 228
> ath:AT5G27140 SAR DNA-binding protein, putative
Length=445
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/149 (41%), Positives = 95/149 (63%), Gaps = 2/149 (1%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGL 62
L VAD L + I ++L+I + + + E++RG++ L+ L++GLDD D +S+ LSH L
Sbjct 57 LAVADPKLGDIITEKLDIECVHNDAVMELLRGVRSQLTELLSGLDDNDLAPVSLELSHIL 116
Query 63 NRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCK 122
R KL+ T DK TMII +I LL++LDKELN + + E YG HFPEL I+ D++LY K
Sbjct 117 ARYKLKITSDK--TMIILSISLLDDLDKELNTYTTSVCELYGLHFPELANIVQDNILYAK 174
Query 123 IIQLIQFRSNAKYAPLETLLPHEICEEIR 151
+++L+ R NA +L E+ E++
Sbjct 175 VVKLMGNRINAATLDFSEILADEVEAELK 203
> dre:100334154 NOP56-like; K14564 nucleolar protein 56
Length=432
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 45/96 (46%), Positives = 67/96 (69%), Gaps = 0/96 (0%)
Query 35 IKLHLSSLITGLDDQDRHRMSMCLSHGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNN 94
++LH SL+ GL Q + + L H +R K++F ++ D MIIQ+I LL++LDK++N
Sbjct 10 VRLHFHSLVKGLTAQAASKAQLGLGHSYSRAKVKFNVNRADNMIIQSIALLDQLDKDINT 69
Query 95 FIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFR 130
F MR++EWYG+HFPEL KI+ D+ YCK+ +LI R
Sbjct 70 FSMRVREWYGYHFPELIKIVSDNSTYCKLAKLIGNR 105
> ath:AT1G56110 NOP56; NOP56 (Arabidopsis homolog of nucleolar
protein Nop56); K14564 nucleolar protein 56
Length=522
Score = 106 bits (265), Expect = 2e-23, Method: Composition-based stats.
Identities = 47/131 (35%), Positives = 83/131 (63%), Gaps = 0/131 (0%)
Query 1 LTLIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSH 60
+L +A+ L ++I + I + E++RG++ H I L D + + L+H
Sbjct 94 FSLGLAEPKLGSHIFEATKIPCQSNEFVLELLRGVRQHFDRFIKDLKPGDLEKSQLGLAH 153
Query 61 GLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLY 120
+R K++F +++D M+IQAI +L+ LDK++N+F MR++EWY WHFPEL KI++D+ LY
Sbjct 154 SYSRAKVKFNVNRVDNMVIQAIFMLDTLDKDINSFAMRVREWYSWHFPELVKIVNDNYLY 213
Query 121 CKIIQLIQFRS 131
++ ++I +S
Sbjct 214 ARVSKMIDDKS 224
> ath:AT3G12860 nucleolar protein Nop56, putative
Length=499
Score = 105 bits (263), Expect = 5e-23, Method: Composition-based stats.
Identities = 43/103 (41%), Positives = 72/103 (69%), Gaps = 0/103 (0%)
Query 29 YEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGLNRLKLRFTPDKLDTMIIQAIGLLEEL 88
+E++RG++ H I L D + + L+H +R K++F +++D M+IQAI +L+ L
Sbjct 122 HELLRGVRQHFDRFIKDLKPGDLEKAQLGLAHSYSRAKVKFNVNRVDNMVIQAIFMLDTL 181
Query 89 DKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRS 131
DK++N+F MR++EWY WHFPEL KI++D+ LY K+ ++I +S
Sbjct 182 DKDINSFAMRVREWYSWHFPELVKIVNDNYLYAKVSKIIVDKS 224
> tpv:TP01_0747 hypothetical protein; K14564 nucleolar protein
56
Length=560
Score = 101 bits (252), Expect = 7e-22, Method: Composition-based stats.
Identities = 53/156 (33%), Positives = 90/156 (57%), Gaps = 8/156 (5%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLH----LSSLITGLDDQDRHRMSMCL 58
L + D +LA ++ + V++ + E++RG +L+ L+ L +G D + + L
Sbjct 90 LAIVDVSLAKSLSHK-GFKVIYDSNVLELVRGCRLYETKRLTKLASGGTSFDMNNFQVGL 148
Query 59 SHGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSL 118
H +R KL+F P K D II ++ LL+ L K LN+F MR++EWYGWHFPEL K++ D+
Sbjct 149 GHSYSRSKLKFDPSKQDKPIINSVSLLDTLTKNLNSFAMRVREWYGWHFPELCKLVPDNK 208
Query 119 LYCKIIQLIQFRSNAKY---APLETLLPHEICEEIR 151
+C+ ++LI+ + + PL LL E+ ++
Sbjct 209 TFCEAVKLIKRKEEFDFDNLEPLNELLGEELALTVK 244
> bbo:BBOV_IV010840 23.m05998; nucleolar protein Nop56; K14564
nucleolar protein 56
Length=569
Score = 101 bits (252), Expect = 8e-22, Method: Composition-based stats.
Identities = 48/138 (34%), Positives = 80/138 (57%), Gaps = 5/138 (3%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLH----LSSLITGLDDQDRHRMSMCL 58
L V D L ++ + VL+ + E++RG + H ++ L +G D + + L
Sbjct 90 LAVVDPALGKSLSAK-GFQVLYDSNVIEILRGCRQHEMKHIAKLASGASAFDMDKFHVGL 148
Query 59 SHGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSL 118
H +R KL+ P + D ++ + LL+ L K LN+F MR++EWYGWHFPEL KI+ D+
Sbjct 149 GHNYSRTKLQVDPRRHDKPVVNCVALLDSLTKNLNSFAMRVREWYGWHFPELVKIVPDNK 208
Query 119 LYCKIIQLIQFRSNAKYA 136
LYC+ +Q+IQ ++ ++
Sbjct 209 LYCQTVQIIQCKNKFDWS 226
> cpv:cgd2_50 SIK1 nucleolar protein Nop56 ; K14564 nucleolar
protein 56
Length=499
Score = 101 bits (252), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 50/127 (39%), Positives = 76/127 (59%), Gaps = 3/127 (2%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLHLSSLITGLDDQ--DRHRMSMCLSH 60
L V D +L + Q V+ + E++RGI++H + ++ D D H+ + L H
Sbjct 96 LGVGDASLGKTLSDQ-GYPVVIDKNINELLRGIRIHFTRIVKSFDSSIGDLHKFQVGLGH 154
Query 61 GLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLY 120
+R KL+F P+K D I+Q+I L++ LDK++N F MR +EWY WHFPEL KII D+ +
Sbjct 155 SFSRNKLQFDPNKQDKSIVQSIALIDRLDKDINLFSMRCREWYSWHFPELAKIITDTEKF 214
Query 121 CKIIQLI 127
K+ LI
Sbjct 215 LKVAVLI 221
> tgo:TGME49_047470 nucleolar protein 5A, putative ; K14564 nucleolar
protein 56
Length=536
Score = 101 bits (251), Expect = 1e-21, Method: Composition-based stats.
Identities = 52/134 (38%), Positives = 81/134 (60%), Gaps = 6/134 (4%)
Query 22 VLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLSHGLNRLKLRFTPDKLDTMIIQA 81
++F+ + E+ RG + H+ L L + + + L H +R K++ P K D I+Q+
Sbjct 128 IVFNSNVQELHRGCRQHMKKLAKQLGELPIEKFQVGLGHSYSRSKMQEDPRKQDKPIMQS 187
Query 82 IGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSN-AKYAPLET 140
I LL+ LDK +N F M++KEWYGWHFPEL KI D+ +YCK+++++Q + ++ E
Sbjct 188 IALLDSLDKNINAFAMKLKEWYGWHFPELVKICGDAEVYCKVLKVVQMKEQFDEHTQGEE 247
Query 141 LLPHEIC---EEIR 151
LL E C EEIR
Sbjct 248 LL--EACGGSEEIR 259
> pfa:PF11_0191 Nop56/Sik1-like protein, putative; K14564 nucleolar
protein 56
Length=594
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 52/141 (36%), Positives = 79/141 (56%), Gaps = 6/141 (4%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLH----LSSLITGLDDQDRHRMSMCL 58
L VAD NL + + N++ + + E+ R + H +S+ + +D +H ++ L
Sbjct 92 LGVADINLGKYLS-NVGFNIIHNNNVLELFRACRQHYLKKISTYVNNIDIDIKH-FNIGL 149
Query 59 SHGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSL 118
H +R KL+ P K D II +IG +E LDK +N F MR+ EWY WHFPEL KI+ D
Sbjct 150 GHSYSRSKLKLDPRKQDKSIINSIGTIESLDKNINLFSMRVIEWYSWHFPELKKIVTDVC 209
Query 119 LYCKIIQLIQFRSNAKYAPLE 139
+YCK++ LIQ + + E
Sbjct 210 MYCKLVNLIQIKEKFDFDTYE 230
> cel:K07C5.4 hypothetical protein; K14564 nucleolar protein 56
Length=486
Score = 88.6 bits (218), Expect = 7e-18, Method: Composition-based stats.
Identities = 40/124 (32%), Positives = 73/124 (58%), Gaps = 1/124 (0%)
Query 1 LTLIVADKNLANNIKKQL-NINVLFSPSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLS 59
+ L + D LA ++ + ++ ++F E++RG ++H L L + + L
Sbjct 93 VVLGINDSKLAGSLTEAFPDLKLVFGGVITEILRGTRVHFERLAKNLPHHSLSKAQLSLG 152
Query 60 HGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLL 119
H +R K++F ++D M+IQ+I LL++LDK++N F MRI+EWY +H+PEL ++ D
Sbjct 153 HSYSRSKVKFDVHRVDNMVIQSIALLDQLDKDINLFGMRIREWYSYHYPELFRLAPDQYK 212
Query 120 YCKI 123
Y ++
Sbjct 213 YSRL 216
> tgo:TGME49_044100 putative snoRNA binding domain-containing
protein ; K12844 U4/U6 small nuclear ribonucleoprotein PRP31
Length=553
Score = 37.4 bits (85), Expect = 0.016, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 87 ELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAKYAPLETLLP 143
++DK++ N IK+ Y FPEL I+ L Y ++ IQ +++ L LLP
Sbjct 138 DIDKDILNIHKFIKDIYSMKFPELESIVQSPLEYIGVVLRIQNQTDLTQVDLSDLLP 194
> pfa:PFD0450c pre-mRNA splicing factor, putative; K12844 U4/U6
small nuclear ribonucleoprotein PRP31
Length=534
Score = 35.0 bits (79), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 75 DTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAK 134
D MI + + + ++D E+ N +K+ Y FPEL+ I++ + Y ++ I+ + K
Sbjct 152 DVMIEKCMETILKIDTEILNIHKYVKDIYSTKFPELDSIVYSPVEYISVVNKIRNEVDLK 211
Query 135 YAPLETLLPH 144
+LP+
Sbjct 212 NIDFSDILPN 221
> tpv:TP02_0595 hypothetical protein; K12844 U4/U6 small nuclear
ribonucleoprotein PRP31
Length=498
Score = 35.0 bits (79), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 87 ELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAKYAPLETLLPHEI 146
++D+E+ N +++ Y FP+L I++ L Y +++ Q S+ L LLP+ +
Sbjct 135 KIDREIINIFNYVRDIYSKRFPKLESIVYSPLDYIAVVKRAQNESDFTKIDLTDLLPNSM 194
> bbo:BBOV_II006320 18.m06519; pre-mRNA processing ribonucleoprotein
binding region-containing protein; K12844 U4/U6 small
nuclear ribonucleoprotein PRP31
Length=483
Score = 34.7 bits (78), Expect = 0.11, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 37/75 (49%), Gaps = 0/75 (0%)
Query 76 TMIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAKY 135
T+I + ++E+D E+ N +++ Y FP+L I++ L Y +++ Q +
Sbjct 93 TLIEECNQAVQEIDNEIINIYNYVRDIYSKRFPKLESIVYSPLDYIAVVRRAQNEMDFTK 152
Query 136 APLETLLPHEICEEI 150
L +LP+ + I
Sbjct 153 VTLSDILPNTMVMAI 167
> xla:779264 rilp, MGC154468; Rab interacting lysosomal protein;
K13883 Rab-interacting lysosomal protein
Length=393
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 26/102 (25%), Positives = 43/102 (42%), Gaps = 13/102 (12%)
Query 49 QDRHRMSMCLSHGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYGWHFP 108
QD + M++CL L L ++ PD + ++ Q + +LE L E Y
Sbjct 15 QDVYNMAVCLGSELQTLTEQYGPDAVSGVVPQVVRVLELL------------ESYAGGSG 62
Query 109 ELNKIIHDSLLYCKIIQLIQFRSNAKYAPLETLLPHEICEEI 150
+ LL + + +QF S + AP E LL + E +
Sbjct 63 RERSCTEEELLI-RAVHRLQFSSREEPAPAEQLLEAQKKEHV 103
> dre:393476 prpf31, MGC66177, zgc:66177; PRP31 pre-mRNA processing
factor 31 homolog (yeast); K12844 U4/U6 small nuclear
ribonucleoprotein PRP31
Length=508
Score = 32.7 bits (73), Expect = 0.46, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 77 MIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQ 125
+I+ A L E+D ELN +++ Y FPEL ++ ++L Y + ++
Sbjct 103 LIVAANNLTVEIDNELNIIHKFVRDKYSKRFPELESLVPNALDYIRTVK 151
> cel:Y110A7A.8 hypothetical protein; K12844 U4/U6 small nuclear
ribonucleoprotein PRP31
Length=504
Score = 32.3 bits (72), Expect = 0.59, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 32/58 (55%), Gaps = 0/58 (0%)
Query 77 MIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAK 134
+I++ + ++D E+N +++ Y FPEL ++ ++L Y +QL+ N+K
Sbjct 101 LIVKLSHVAADIDNEINVIHKFVRDKYEKRFPELETLVPNALTYLATVQLLGNEINSK 158
> mmu:68988 Prpf31, 1500019O16Rik, 2810404O06Rik, AW554706, PRP31,
RP11; PRP31 pre-mRNA processing factor 31 homolog (yeast);
K12844 U4/U6 small nuclear ribonucleoprotein PRP31
Length=493
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 77 MIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQ 125
+I+ A L E++ ELN I++ Y FPEL ++ ++L Y + ++
Sbjct 92 VIVDANNLTVEIENELNIIHKFIRDKYSKRFPELESLVPNALDYIRTVK 140
> hsa:26121 PRPF31, DKFZp566J153, NY-BR-99, PRP31, RP11; PRP31
pre-mRNA processing factor 31 homolog (S. cerevisiae); K12844
U4/U6 small nuclear ribonucleoprotein PRP31
Length=499
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 77 MIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQ 125
+I+ A L E++ ELN I++ Y FPEL ++ ++L Y + ++
Sbjct 92 VIVDANNLTVEIENELNIIHKFIRDKYSKRFPELESLVPNALDYIRTVK 140
> xla:495301 prpf31, prp31, rp11; PRP31 pre-mRNA processing factor
31 homolog; K12844 U4/U6 small nuclear ribonucleoprotein
PRP31
Length=498
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 77 MIIQAIGLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQ 125
+I+ A L E++ ELN I++ Y FPEL ++ ++L Y + ++
Sbjct 91 VIVDANNLTVEIENELNIIHKFIRDKYSKRFPELESLVPNALDYIRTVK 139
> xla:378573 slc13a2, NaDC-2, nact, nadc-1, nadc1, nadc2-a, sdct1;
solute carrier family 13 (sodium-dependent dicarboxylate
transporter), member 2; K14445 solute carrier family 13 (sodium-dependent
dicarboxylate transporter), member 2/3/5
Length=622
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query 83 GLLEELDKELNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQL-IQFRSN 132
G ++EL E NN I+ W+G+ FP + ++ S L+ + I L + F+ N
Sbjct 287 GQMDELFPE-NNNIINFASWFGFAFPTMLVLLALSWLWLQFIYLGVNFKKN 336
> cel:R07B7.14 nhr-207; Nuclear Hormone Receptor family member
(nhr-207)
Length=406
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/59 (25%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 46 LDDQDRHRMSMCLSHGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIKEWYG 104
L+D+ R + L++ L + +R+ PD+L + I+ + ++E KE +F + ++ +Y
Sbjct 333 LEDERRRYVRSLLTYCLKQYGIRYGPDRLSS-ILSIMPIMENQQKEEKSFNVILRSFYS 390
> xla:379261 MGC53392; similar to angiopoietin-like factor
Length=343
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/101 (19%), Positives = 53/101 (52%), Gaps = 3/101 (2%)
Query 1 LTLIVADKNLANNIKKQLNINVLFS-PSCYEVIRGIKLHLSSLITGLDDQDRHRMSMCLS 59
+L + L K++ N +F P+C E ++ +K+ +S+L + L + + + + ++
Sbjct 14 FSLAASPATLQKVPKRKPGANGIFKVPACCEELKDLKVQISNLSSLLGELSKKQETDWVN 73
Query 60 HGLNRLKLRFTPDKLDTMIIQAIGLLEELDKELNNFIMRIK 100
+ +KL + K++T + G E++ +++ IM+++
Sbjct 74 VVMQVMKLEGSYKKVETRVTDVEGKYSEMNNQMD--IMQLQ 112
> mmu:20745 Spock1, Spock, Ticn1, testican; sparc/osteonectin,
cwcv and kazal-like domains proteoglycan 1; K08136 sparc/osteonectin,
cwcv and kazal-like domains proteoglycan (testican)
Length=439
Score = 28.9 bits (63), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 89 DKELNNFIMRIKEWYGWHFPELNKII 114
DKEL N R+K+W+G + N++I
Sbjct 200 DKELRNLASRLKDWFGALHEDANRVI 225
> hsa:6695 SPOCK1, FLJ37170, SPOCK, TESTICAN, TIC1; sparc/osteonectin,
cwcv and kazal-like domains proteoglycan (testican)
1; K08136 sparc/osteonectin, cwcv and kazal-like domains proteoglycan
(testican)
Length=439
Score = 28.5 bits (62), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 89 DKELNNFIMRIKEWYGWHFPELNKII 114
DKEL N R+K+W+G + N++I
Sbjct 200 DKELRNLASRLKDWFGALHEDANRVI 225
> ath:AT1G50310 STP9; STP9 (SUGAR TRANSPORTER 9); carbohydrate
transmembrane transporter/ sugar:hydrogen symporter
Length=517
Score = 28.5 bits (62), Expect = 8.2, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 37/65 (56%), Gaps = 5/65 (7%)
Query 76 TMIIQAIG-LLEELDKELNNFIMRIKEWYGWHFPELNKIIHDS---LLYCKII-QLIQFR 130
T I+ A+G LL D ++ + ++E+ FPE++K +H++ YCK QL+Q
Sbjct 28 TCIVAAMGGLLFGYDLGISGGVTSMEEFLSKFFPEVDKQMHEARRETAYCKFDNQLLQLF 87
Query 131 SNAKY 135
+++ Y
Sbjct 88 TSSLY 92
> sce:YGR091W PRP31; Prp31p; K12844 U4/U6 small nuclear ribonucleoprotein
PRP31
Length=494
Score = 28.5 bits (62), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 6/49 (12%)
Query 92 LNNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAKYAPLET 140
++NF++ + Y FPEL+ +I L Y K+I +++ N Y+ E+
Sbjct 112 MHNFLISL---YSRRFPELSSLIPSPLQYSKVISILE---NENYSKNES 154
> sce:YPL269W KAR9; Kar9p
Length=644
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 17/109 (15%)
Query 50 DRHRMSMCLSHGLNRLKLRFTPDKLDT-----------MIIQAIGLLEELD------KEL 92
D H + +H L+ L+ F+ D D+ +I ++ + +L+ KEL
Sbjct 99 DVHNIEEGFAHLLDLLEDEFSKDDQDSDKYNRFSPMFDVIEESTQIKTQLEPWLTNLKEL 158
Query 93 NNFIMRIKEWYGWHFPELNKIIHDSLLYCKIIQLIQFRSNAKYAPLETL 141
+ + E H L+KII+ ++ YC IQ +F S ++ P TL
Sbjct 159 LDTSLEFNEISKDHMDTLHKIINSNISYCLEIQEERFASPIRHTPSFTL 207
> cel:F15B9.6 hypothetical protein
Length=458
Score = 28.5 bits (62), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 33/148 (22%), Positives = 63/148 (42%), Gaps = 25/148 (16%)
Query 3 LIVADKNLANNIKKQLNINVLFSPSCYEVIRGIKLH--LSSLITGLDDQDRHRMSMCLSH 60
+ D NL +K +N + S + + I+ H +++ + L D D ++
Sbjct 285 FVTFDGNLHAAMKPMGTLNSILSAT----VHKIQPHPVINATLRHLFDNDVYKYGWSNRI 340
Query 61 GLNRLKLRF----TPDKLDTMIIQAIGLLEELDKELNNF-------------IMRIKE-- 101
++ LRF T DK+D +IQ G + +D E +F +++I E
Sbjct 341 EIDSCILRFEHCKTLDKIDYCLIQLRGCILSIDNEDADFKSTTKTIYATIDELIKIPEES 400
Query 102 WYGWHFPELNKIIHDSLLYCKIIQLIQF 129
W+ P + II + +C I + ++F
Sbjct 401 WWSTILPVIAIIISIFVCFCDIFRCVKF 428
Lambda K H
0.325 0.142 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40