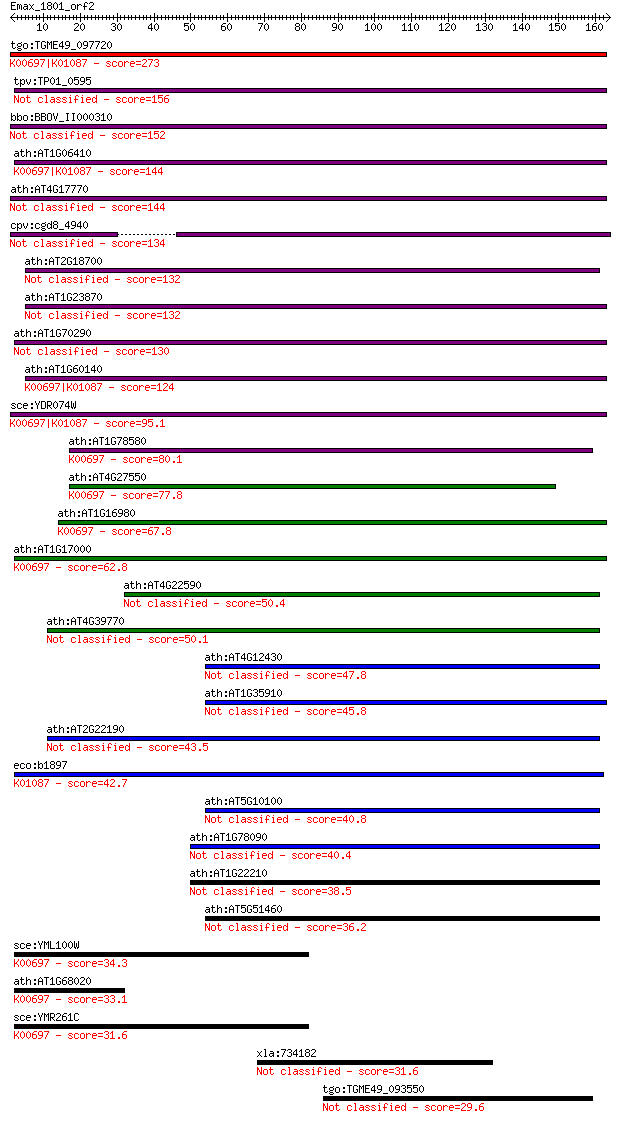
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1801_orf2
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_097720 trehalose-6-phosphate synthase domain-contai... 273 2e-73
tpv:TP01_0595 trehalose-6-phosphate synthase 156 3e-38
bbo:BBOV_II000310 18.m06009; trehalose-6-phosphate synthase do... 152 6e-37
ath:AT1G06410 ATTPS7; ATTPS7; alpha,alpha-trehalose-phosphate ... 144 9e-35
ath:AT4G17770 ATTPS5; ATTPS5; protein binding / transferase, t... 144 2e-34
cpv:cgd8_4940 trehalose-6-phosphate synthase of likely plant o... 134 1e-31
ath:AT2G18700 ATTPS11; transferase, transferring glycosyl groups 132 3e-31
ath:AT1G23870 ATTPS9; ATTPS9; transferase, transferring glycos... 132 6e-31
ath:AT1G70290 ATTPS8; ATTPS8; alpha,alpha-trehalose-phosphate ... 130 2e-30
ath:AT1G60140 ATTPS10 (trehalose phosphate synthase); transfer... 124 2e-28
sce:YDR074W TPS2, HOG2, PFK3; Phosphatase subunit of the treha... 95.1 9e-20
ath:AT1G78580 ATTPS1; ATTPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE);... 80.1 3e-15
ath:AT4G27550 ATTPS4; ATTPS4; alpha,alpha-trehalose-phosphate ... 77.8 2e-14
ath:AT1G16980 ATTPS2; ATTPS2; alpha,alpha-trehalose-phosphate ... 67.8 1e-11
ath:AT1G17000 ATTPS3; ATTPS3; alpha,alpha-trehalose-phosphate ... 62.8 5e-10
ath:AT4G22590 trehalose-6-phosphate phosphatase, putative (EC:... 50.4 3e-06
ath:AT4G39770 trehalose-6-phosphate phosphatase, putative (EC:... 50.1 3e-06
ath:AT4G12430 trehalose-6-phosphate phosphatase, putative (EC:... 47.8 2e-05
ath:AT1G35910 trehalose-6-phosphate phosphatase, putative 45.8 7e-05
ath:AT2G22190 catalytic/ trehalose-phosphatase (EC:3.1.3.12) 43.5 3e-04
eco:b1897 otsB, ECK1896, JW1886, otsP; trehalose-6-phosphate p... 42.7 6e-04
ath:AT5G10100 trehalose-6-phosphate phosphatase, putative (EC:... 40.8 0.002
ath:AT1G78090 ATTPPB; ATTPPB (TREHALOSE-6-PHOSPHATE PHOSPHATAS... 40.4 0.003
ath:AT1G22210 trehalose-6-phosphate phosphatase, putative 38.5 0.009
ath:AT5G51460 ATTPPA; ATTPPA; trehalose-phosphatase (EC:3.1.3.12) 36.2 0.045
sce:YML100W TSL1; Large subunit of trehalose 6-phosphate synth... 34.3 0.20
ath:AT1G68020 ATTPS6; ATTPS6; alpha,alpha-trehalose-phosphate ... 33.1 0.44
sce:YMR261C TPS3; Regulatory subunit of trehalose-6-phosphate ... 31.6 1.1
xla:734182 ara-55; Hic-5 31.6 1.4
tgo:TGME49_093550 hypothetical protein 29.6 4.4
> tgo:TGME49_097720 trehalose-6-phosphate synthase domain-containing
protein (EC:2.4.1.15 2.7.9.5 3.1.3.12); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15];
K01087 trehalose-phosphatase [EC:3.1.3.12]
Length=1222
Score = 273 bits (699), Expect = 2e-73, Method: Compositional matrix adjust.
Identities = 124/162 (76%), Positives = 143/162 (88%), Gaps = 0/162 (0%)
Query 1 DRSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKR 60
DR LLE+WF ++GIGLCAEHG+YYR+ +TGD+W CM DFTWK VA+ELMLQYVKR
Sbjct 833 DRHLLEEWFSSIRGIGLCAEHGFYYRVPGITGDQWHCMSRQTDFTWKQVAIELMLQYVKR 892
Query 61 TQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLG 120
TQGSF ENKGSALVFQYRDADPDFG +QAKDLS++L ELLFGYPV+V+SGKGYVEVKL G
Sbjct 893 TQGSFIENKGSALVFQYRDADPDFGSMQAKDLSNYLGELLFGYPVSVMSGKGYVEVKLRG 952
Query 121 VNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
VNKG AVE++L+ LS LHGD+DFV C+GDDRSDEDMFAVIN+
Sbjct 953 VNKGHAVEKVLRKLSNLHGDVDFVLCVGDDRSDEDMFAVINA 994
> tpv:TP01_0595 trehalose-6-phosphate synthase
Length=1059
Score = 156 bits (395), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 76/181 (41%), Positives = 114/181 (62%), Gaps = 20/181 (11%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGC-------------------MCPDA 42
+ L++ WF + +GLCAE GYY++L + G EW + PD
Sbjct 815 KELMDSWFSSIPDVGLCAERGYYFKLPTVLGREWQTFRHLSTNATSNHTNSAQLEVEPDV 874
Query 43 DF-TWKSVAVELMLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLF 101
+W+ VA++L QYV+RT GS+ E+K S LVFQ++++D +FG LQA +L+S L EL+
Sbjct 875 PSDSWRPVALKLFEQYVQRTPGSYLESKDSCLVFQFQNSDQEFGILQANELNSSLCELMG 934
Query 102 GYPVTVVSGKGYVEVKLLGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVIN 161
G+PV V GKGY+E++L G+NKG A+ ++ S+L GD DFV C+GDD+SDE+ F +
Sbjct 935 GFPVDVHRGKGYLELRLKGINKGNALVNVVHKYSSLFGDFDFVLCLGDDKSDEEAFKALE 994
Query 162 S 162
+
Sbjct 995 T 995
> bbo:BBOV_II000310 18.m06009; trehalose-6-phosphate synthase
domain containing protein
Length=958
Score = 152 bits (383), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 69/162 (42%), Positives = 101/162 (62%), Gaps = 0/162 (0%)
Query 1 DRSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKR 60
+R LE WF D+ +GLCA +GY+Y++ LTG +W CM + D WK +A+++M QY R
Sbjct 740 NRDCLETWFPDLPNLGLCAGYGYFYKIPFLTGGKWNCMVENVDDRWKVIALQIMEQYAHR 799
Query 61 TQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLG 120
T GS+ EN +VFQY +DP+F Q+ +L + L +++ YPV V K V V L G
Sbjct 800 TPGSYIENMDVMVVFQYHHSDPEFSATQSVELLTVLKQVMAPYPVDVQRTKWNVHVCLRG 859
Query 121 VNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
VNKG + I + ++GD DF+ C+GD RSDE+MF + +
Sbjct 860 VNKGATLLNIAENYCRIYGDFDFILCVGDHRSDEEMFKALET 901
> ath:AT1G06410 ATTPS7; ATTPS7; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]; K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=851
Score = 144 bits (364), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 76/161 (47%), Positives = 95/161 (59%), Gaps = 2/161 (1%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRT 61
R L WF K IG+ AEHGY+ + +EW +DF W + +M QY + T
Sbjct 633 RESLSKWFTPCKKIGIAAEHGYFLKWS--GSEEWETCGQSSDFGWMQIVEPVMKQYTEST 690
Query 62 QGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGV 121
GS E K SALV+QYRDADP FG LQAK++ HL +L PV V SG VEVK GV
Sbjct 691 DGSSIEIKESALVWQYRDADPGFGSLQAKEMLEHLESVLANEPVAVKSGHYIVEVKPQGV 750
Query 122 NKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
+KG E+I +++ +DFV CIGDDRSDEDMF I +
Sbjct 751 SKGSVSEKIFSSMAGKGKPVDFVLCIGDDRSDEDMFEAIGN 791
> ath:AT4G17770 ATTPS5; ATTPS5; protein binding / transferase,
transferring glycosyl groups / trehalose-phosphatase
Length=862
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 76/162 (46%), Positives = 98/162 (60%), Gaps = 2/162 (1%)
Query 1 DRSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKR 60
DR L +WF +GL AEHGY+ R + G +W + F WK +A +M Y +
Sbjct 638 DRRTLTEWFSSCDDLGLGAEHGYFIRPND--GTDWETSSLVSGFEWKQIAEPVMRLYTET 695
Query 61 TQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLG 120
T GS E K +ALV+ Y+ ADPDFG QAK+L HL +L PV+V +G+ VEVK G
Sbjct 696 TDGSTIETKETALVWNYQFADPDFGSCQAKELMEHLESVLTNDPVSVKTGQQLVEVKPQG 755
Query 121 VNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
VNKG ER+L T+ +DF+ C+GDDRSDEDMF VI S
Sbjct 756 VNKGLVAERLLTTMQEKGKLLDFILCVGDDRSDEDMFEVIMS 797
> cpv:cgd8_4940 trehalose-6-phosphate synthase of likely plant
origin
Length=1417
Score = 134 bits (337), Expect = 1e-31, Method: Composition-based stats.
Identities = 66/150 (44%), Positives = 93/150 (62%), Gaps = 32/150 (21%)
Query 46 WKSVAVELMLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPV 105
WK++ +LM QYV RTQGS+ ENKG+ALVFQ++ +P FG QAK+LS++LSELL +PV
Sbjct 1212 WKNITFQLMEQYVLRTQGSYIENKGTALVFQFKYCEPYFGAWQAKELSNYLSELLINFPV 1271
Query 106 TVVSGKGYVEVKLLGVNKGKAVERILQTLSTLHGD------------------------- 140
V+SG +VEV+L G+NKG AV+ IL +++L +
Sbjct 1272 NVISGNDFVEVRLQGINKGVAVQAILNQINSLSRNNNNYENKSEEDDLTKNNASPNLSSS 1331
Query 141 -------IDFVFCIGDDRSDEDMFAVINSW 163
++ + CIGDDRSDEDMF+V+N +
Sbjct 1332 FHSPPPPLEMILCIGDDRSDEDMFSVVNHF 1361
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 18/29 (62%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 1 DRSLLEDWFRDVKGIGLCAEHGYYYRLDK 29
+R LL++WF D+ IGLCAEHGYY +L K
Sbjct 1039 ERHLLDEWFGDIDNIGLCAEHGYYLKLPK 1067
> ath:AT2G18700 ATTPS11; transferase, transferring glycosyl groups
Length=862
Score = 132 bits (333), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 70/157 (44%), Positives = 93/157 (59%), Gaps = 3/157 (1%)
Query 5 LEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRTQGS 64
L WF +G+ AEHGY+ R + + W AD +WK +A +M Y++ T GS
Sbjct 634 LSKWFDSCPNLGISAEHGYFTRWN--SNSPWETSELPADLSWKKIAKPVMNHYMEATDGS 691
Query 65 FTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKG 124
F E K SA+V+ +++AD FG QAK+L HL +L PV V G+ VEVK GV+KG
Sbjct 692 FIEEKESAMVWHHQEADHSFGSWQAKELLDHLESVLTNEPVVVKRGQHIVEVKPQGVSKG 751
Query 125 KAVERILQTLSTLHGD-IDFVFCIGDDRSDEDMFAVI 160
K VE ++ T+ G DF+ CIGDDRSDEDMF I
Sbjct 752 KVVEHLIATMRNTKGKRPDFLLCIGDDRSDEDMFDSI 788
> ath:AT1G23870 ATTPS9; ATTPS9; transferase, transferring glycosyl
groups / trehalose-phosphatase
Length=867
Score = 132 bits (331), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 71/158 (44%), Positives = 92/158 (58%), Gaps = 2/158 (1%)
Query 5 LEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRTQGS 64
L DW + +G+ AEHGY+ R + EW A+ WK++ +M Y+ T GS
Sbjct 642 LSDWLSPCENLGIAAEHGYFIRWS--SKKEWETCYSSAEAEWKTMVEPVMRSYMDATDGS 699
Query 65 FTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKG 124
E K SALV+ ++DADPDFG QAK+L HL +L PV V G+ VEVK GV+KG
Sbjct 700 TIEYKESALVWHHQDADPDFGACQAKELLDHLESVLANEPVVVKRGQHIVEVKPQGVSKG 759
Query 125 KAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
AVE+++ + D V CIGDDRSDEDMF I S
Sbjct 760 LAVEKVIHQMVEDGNPPDMVMCIGDDRSDEDMFESILS 797
> ath:AT1G70290 ATTPS8; ATTPS8; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase
Length=856
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 68/161 (42%), Positives = 94/161 (58%), Gaps = 2/161 (1%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRT 61
R L +W + +G+ AEHGY+ R + DEW D W+S+ +M Y++ T
Sbjct 634 RESLSNWLSPCENLGIAAEHGYFIRWK--SKDEWETCYSPTDTEWRSMVEPVMRSYMEAT 691
Query 62 QGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGV 121
G+ E K SALV+ ++DADPDFG QAK++ HL +L PV V G+ VEVK GV
Sbjct 692 DGTSIEFKESALVWHHQDADPDFGSCQAKEMLDHLESVLANEPVVVKRGQHIVEVKPQGV 751
Query 122 NKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
+KG A E++++ + + V CIGDDRSDEDMF I S
Sbjct 752 SKGLAAEKVIREMVERGEPPEMVMCIGDDRSDEDMFESILS 792
> ath:AT1G60140 ATTPS10 (trehalose phosphate synthase); transferase,
transferring glycosyl groups / trehalose-phosphatase;
K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]; K01087 trehalose-phosphatase [EC:3.1.3.12]
Length=861
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 69/158 (43%), Positives = 89/158 (56%), Gaps = 2/158 (1%)
Query 5 LEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRTQGS 64
L +W + +G+ AEHGY+ R +K +W D WK V +M Y + T GS
Sbjct 642 LSEWLAPCENLGIAAEHGYFTRWNK--SSDWETSGLSDDLEWKKVVEPIMRLYTETTDGS 699
Query 65 FTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKG 124
E K SALV+ ++DADPDFG QAK+L HL +L PV V G VEVK GV+KG
Sbjct 700 NIEAKESALVWHHQDADPDFGSCQAKELLDHLETVLVNEPVIVNRGHQIVEVKPQGVSKG 759
Query 125 KAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
+IL + DFV CIGDDRSDE+MF I++
Sbjct 760 LVTGKILSRMLEDGIAPDFVVCIGDDRSDEEMFENIST 797
> sce:YDR074W TPS2, HOG2, PFK3; Phosphatase subunit of the trehalose-6-phosphate
synthase/phosphatase complex, which synthesizes
the storage carbohydrate trehalose; expression is induced
by stress conditions and repressed by the Ras-cAMP pathway
(EC:3.1.3.12); K00697 alpha,alpha-trehalose-phosphate synthase
(UDP-forming) [EC:2.4.1.15]; K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=896
Score = 95.1 bits (235), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 56/177 (31%), Positives = 88/177 (49%), Gaps = 17/177 (9%)
Query 1 DRSLLEDWFR-DVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVK 59
D+ L W + +GL AEHG + + ++ +W + D +W+ E+M ++
Sbjct 620 DQKFLNKWLGGKLPQLGLSAEHGCF--MKDVSCQDWVNLTEKVDMSWQVRVNEVMEEFTT 677
Query 60 RTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLL 119
RT GSF E K AL + YR P+ G+ AK+L L + + V+ GK +EV+
Sbjct 678 RTPGSFIERKKVALTWHYRRTVPELGEFHAKELKEKLLSFTDDFDLEVMDGKANIEVRPR 737
Query 120 GVNKGKAVERILQTLSTLHGDI--------------DFVFCIGDDRSDEDMFAVINS 162
VNKG+ V+R++ D+ DFV C+GDD +DEDMF +N+
Sbjct 738 FVNKGEIVKRLVWHQHGKPQDMLKGISEKLPKDEMPDFVLCLGDDFTDEDMFRQLNT 794
> ath:AT1G78580 ATTPS1; ATTPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE);
alpha,alpha-trehalose-phosphate synthase (UDP-forming)/ transferase,
transferring glycosyl groups; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=942
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 52/148 (35%), Positives = 80/148 (54%), Gaps = 9/148 (6%)
Query 17 LCAEHGYYYRLDKLTGDEWGCMCPDA-DFTWKSVAVELMLQYVKRTQGSFTENKGSALVF 75
L AE+G + RL T EW P+ + W + + +RT S E + ++L++
Sbjct 662 LAAENGMFLRL---TNGEWMTTMPEHLNMEWVDSVKHVFKYFTERTPRSHFETRDTSLIW 718
Query 76 QYRDADPDFGQLQAKDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGKAVERILQTL 134
Y+ AD +FG+LQA+DL HL + + V VV G VEV+ +GV KG A++RIL +
Sbjct 719 NYKYADIEFGRLQARDLLQHLWTGPISNASVDVVQGSRSVEVRAVGVTKGAAIDRILGEI 778
Query 135 ---STLHGDIDFVFCIGDDR-SDEDMFA 158
++ ID+V CIG DED++
Sbjct 779 VHSKSMTTPIDYVLCIGHFLGKDEDVYT 806
> ath:AT4G27550 ATTPS4; ATTPS4; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups; K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=795
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 49/137 (35%), Positives = 73/137 (53%), Gaps = 8/137 (5%)
Query 17 LCAEHGYYYRLDKLTGDEWGCMCPD-ADFTWKSVAVELMLQYVKRTQGSFTENKGSALVF 75
L AE+G + R T EW P+ + W + + +RT GS+ E ++LV+
Sbjct 580 LAAENGMFLRH---TSGEWVTRIPEHMNLEWIDGVKHVFKYFTERTPGSYLETSEASLVW 636
Query 76 QYRDADPDFGQLQAKDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGKAVERILQTL 134
Y +AD +FG+ QA+D+ HL + + V VV G VEV +GV KG A+ERIL +
Sbjct 637 NYENADAEFGRAQARDMLQHLWAGPISNASVDVVRGGQSVEVHAVGVTKGSAMERILGEI 696
Query 135 ---STLHGDIDFVFCIG 148
++ ID+V CIG
Sbjct 697 VHNKSMATPIDYVLCIG 713
> ath:AT1G16980 ATTPS2; ATTPS2; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups; K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=821
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 52/155 (33%), Positives = 76/155 (49%), Gaps = 9/155 (5%)
Query 14 GIGLCAEHGYYYRLDKLTGDEWGCMCP-DADFTWKSVAVELMLQYVKRTQGSFTENKGSA 72
I L AE+G + +K T EW P + + W + + RT S+ E ++
Sbjct 572 NIWLAAENGMF---EKQTTGEWVTNMPQNVNLDWVDGVKNVFKYFTDRTPRSYFEASETS 628
Query 73 LVFQYRDADPDFGQLQAKDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGKAVERIL 131
LV+ Y AD +FG+ QA+DL +L + + V VV G VEV +G KG A+ RIL
Sbjct 629 LVWNYEYADVEFGRAQARDLLQYLWAGPISNASVDVVRGNHSVEVHAIGETKGAAIGRIL 688
Query 132 QTL---STLHGDIDFVFCIGD-DRSDEDMFAVINS 162
+ ++ IDFVFC G DED++ S
Sbjct 689 GEIVHRKSMTTPIDFVFCSGYFLEKDEDIYTFFES 723
> ath:AT1G17000 ATTPS3; ATTPS3; alpha,alpha-trehalose-phosphate
synthase (UDP-forming); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=783
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 53/167 (31%), Positives = 81/167 (48%), Gaps = 10/167 (5%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCP-DADFTWKSVAVELMLQYVKR 60
+++L+ F + K I L AE+G + K T +EW P + + W + + R
Sbjct 539 KNILDKNFGEYK-IWLAAENGMFL---KHTTEEWVTNMPQNMNLDWVDGLKNVFKYFTDR 594
Query 61 TQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHL-SELLFGYPVTVVSGKGYVEVKLL 119
T SF E ++LV+ Y AD +FG+ QA+DL +L + + VV GK VEV +
Sbjct 595 TPRSFFEASKTSLVWNYEYADVEFGRAQARDLLQYLWAGPISNASAEVVRGKYSVEVHAI 654
Query 120 GVNKGKAVERILQTL---STLHGDIDFVFCIGD-DRSDEDMFAVINS 162
GV K + IL + + ID+VFC G DED++ S
Sbjct 655 GVTKEPEIGHILGEIVHKKAMTTPIDYVFCSGYFLEKDEDIYTFFES 701
> ath:AT4G22590 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=377
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 66/134 (49%), Gaps = 11/134 (8%)
Query 32 GDEWGCMCPDADFTWKSVAVEL---MLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQ 88
G+E P +F V E+ +++ K +G+ EN YR+ D L
Sbjct 200 GEEVNLFQPAKEFI--PVIEEVYNNLVEITKCIKGAKVENHKFCTSVHYRNVDEKDWPLV 257
Query 89 AKDLSSHLSELLFGYP-VTVVSGKGYVEVK-LLGVNKGKAVERILQTLSTLHGDIDFVFC 146
A+ + HL YP + + G+ +EV+ ++ NKGKAVE +L++L + D
Sbjct 258 AQRVHDHLKR----YPRLRITHGRKVLEVRPVIEWNKGKAVEFLLESLGLSNNDEFLPIF 313
Query 147 IGDDRSDEDMFAVI 160
IGDD++DED F V+
Sbjct 314 IGDDKTDEDAFKVL 327
> ath:AT4G39770 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=349
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 76/154 (49%), Gaps = 14/154 (9%)
Query 11 DVKGIGLCAEHGYYYRLDKLTGDEWGCMC-PDADFTWKSVAV-ELMLQYVKRTQGSFTEN 68
D+KG E G Y +++ D +C P +F V +++ K T G+ EN
Sbjct 159 DIKG----PEQGSKY--EQILQDSKSLLCQPATEFLPMIDEVYHKLVEKTKSTPGAQVEN 212
Query 69 KGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKGYVEVK-LLGVNKGKA 126
+ +R D + DL++ + ++ YP + + G+ +EV+ ++ +KGKA
Sbjct 213 NKFCVSVHFRRVDEN----NWSDLANQVRSVMKDYPKLRLTQGRKVLEVRPIIKWDKGKA 268
Query 127 VERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
+E +L++L + F IGDDR+DED F V+
Sbjct 269 LEFLLESLGYANCTDVFPLYIGDDRTDEDAFKVL 302
> ath:AT4G12430 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=368
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 58/109 (53%), Gaps = 6/109 (5%)
Query 54 MLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 112
+++ +K +G+ EN YR+ D + A+ + HL + YP + + G+
Sbjct 214 LVEKMKDIKGAKVENHKFCASVHYRNVDEKDWPIIAQRVHDHLKQ----YPRLRLTHGRK 269
Query 113 YVEVK-LLGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
+EV+ ++ NKG+AVE +L++L + D IGDD +DED F V+
Sbjct 270 VLEVRPVIDWNKGRAVEFLLESLGLSNKDDLLPIYIGDDTTDEDAFKVL 318
> ath:AT1G35910 trehalose-6-phosphate phosphatase, putative
Length=369
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 61/111 (54%), Gaps = 6/111 (5%)
Query 54 MLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 112
+++ ++ +G+ EN + YR D L A+ ++S LSE YP + + G+
Sbjct 217 LVEKMRDIEGANVENNKFCVSVHYRCVDQKDWGLVAEHVTSILSE----YPKLRLTQGRK 272
Query 113 YVEVK-LLGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVINS 162
+E++ + +KGKA+E +L++L + + IGDDR+DED F V+ +
Sbjct 273 VLEIRPTIKWDKGKALEFLLESLGFANSNDVLPIYIGDDRTDEDAFKVLRN 323
> ath:AT2G22190 catalytic/ trehalose-phosphatase (EC:3.1.3.12)
Length=354
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 42/156 (26%), Positives = 75/156 (48%), Gaps = 21/156 (13%)
Query 11 DVKGIGLCAEHGYYYRLDKLTGDEWGCMC-PDADFTWKSVAVELMLQYVKRTQ---GSFT 66
D+KG E G Y+ + +C P +F V E+ + V+ TQ G+
Sbjct 167 DIKG----PEQGSKYK-----KENQSLLCQPATEFL--PVINEVYKKLVENTQSIPGAKV 215
Query 67 ENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKGYVEVK-LLGVNKG 124
EN +R + + + DL+ + +L YP + + G+ +E++ ++ +KG
Sbjct 216 ENNKFCASVHFRCVEEN----KWSDLAHQVRSVLKNYPKLMLTQGRKVLEIRPIIKWDKG 271
Query 125 KAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
KA+E +L++L + F IGDDR+DED F ++
Sbjct 272 KALEFLLESLGYDNCTDVFPIYIGDDRTDEDAFKIL 307
> eco:b1897 otsB, ECK1896, JW1886, otsP; trehalose-6-phosphate
phosphatase, biosynthetic (EC:3.1.3.12); K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=266
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 45/162 (27%), Positives = 72/162 (44%), Gaps = 15/162 (9%)
Query 2 RSLLE-DWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKR 60
RS++E D L HG R + G PDA + ++V+L + +
Sbjct 62 RSMVELDALAKPYRFPLAGVHGAERR--DINGKTHIVHLPDA--IARDISVQLH-TVIAQ 116
Query 61 TQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKGYVEVKLL 119
G+ E KG A YR A L L+ ++++ +P + + GK VE+K
Sbjct 117 YPGAELEAKGMAFALHYRQAPQHEDALMT--LAQRITQI---WPQMALQQGKCVVEIKPR 171
Query 120 GVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVIN 161
G +KG+A+ +Q + F +GDD +DE FAV+N
Sbjct 172 GTSKGEAIAAFMQEAPFIGRTPVF---LGDDLTDESGFAVVN 210
> ath:AT5G10100 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=369
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 57/109 (52%), Gaps = 6/109 (5%)
Query 54 MLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 112
+L+ K T G+ EN +R D + + +L + +L +P + + G+
Sbjct 218 LLEKTKSTPGAKVENHKFCASVHFRCVD----EKKWSELVLQVRSVLKKFPTLQLTQGRK 273
Query 113 YVEVK-LLGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
E++ ++ +KGKA+E +L++L + + F IGDDR+DED F ++
Sbjct 274 VFEIRPMIEWDKGKALEFLLESLGFGNTNNVFPVYIGDDRTDEDAFKML 322
> ath:AT1G78090 ATTPPB; ATTPPB (TREHALOSE-6-PHOSPHATE PHOSPHATASE);
trehalose-phosphatase
Length=374
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 56/113 (49%), Gaps = 6/113 (5%)
Query 50 AVELMLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVV 108
V ++ + K G+ EN L +R D + + L+ + +L YP + +
Sbjct 218 VVNILEEKTKWIPGAMVENNKFCLSVHFRRVD----EKRWPALAEVVKSVLIDYPKLKLT 273
Query 109 SGKGYVEVK-LLGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
G+ +E++ + +KG+A+ +L++L + D IGDDR+DED F V+
Sbjct 274 QGRKVLEIRPTIKWDKGQALNFLLKSLGYENSDDVVPVYIGDDRTDEDAFKVL 326
> ath:AT1G22210 trehalose-6-phosphate phosphatase, putative
Length=320
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 53/113 (46%), Gaps = 6/113 (5%)
Query 50 AVELMLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVV 108
V ++ + K G+ E+ L +R D + L+ + +L YP + +
Sbjct 165 VVNVLKEKTKSIPGATVEHNKFCLTVHFRRVD----ETGWAALAEQVRLVLIDYPKLRLT 220
Query 109 SGKGYVEVKL-LGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
G+ +E++ + +KGKA+E +L +L IGDDR+DED F V+
Sbjct 221 QGRKVLELRPSIKWDKGKALEFLLNSLGIAESKDVLPVYIGDDRTDEDAFKVL 273
> ath:AT5G51460 ATTPPA; ATTPPA; trehalose-phosphatase (EC:3.1.3.12)
Length=384
Score = 36.2 bits (82), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 57/109 (52%), Gaps = 6/109 (5%)
Query 54 MLQYVKRTQGSFTENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 112
+++ K +G E+ + YR+ + L A+ + +++ YP + + G+
Sbjct 234 LIESTKDIKGVKVEDNKFCISVHYRNVEEKNWTLVAQ----CVDDVIRTYPKLRLTHGRK 289
Query 113 YVEVK-LLGVNKGKAVERILQTLSTLHGDIDFVFCIGDDRSDEDMFAVI 160
+E++ ++ +KGKAV +L++L + + +GDDR+DED F V+
Sbjct 290 VLEIRPVIDWDKGKAVTFLLESLGLNNCEDVLPIYVGDDRTDEDAFKVL 338
> sce:YML100W TSL1; Large subunit of trehalose 6-phosphate synthase
(Tps1p)/phosphatase (Tps2p) complex, which converts uridine-5'-diphosphoglucose
and glucose 6-phosphate to trehalose,
homologous to Tps3p and may share function; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=1098
Score = 34.3 bits (77), Expect = 0.20, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 41/80 (51%), Gaps = 6/80 (7%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRT 61
+ +LE+ + V+ IGL AE+G Y L+ + W + D W++ +++ V+R
Sbjct 864 KPILENLYSRVQNIGLIAENGAYVSLNGV----WYNIVDQVD--WRNDVAKILEDKVERL 917
Query 62 QGSFTENKGSALVFQYRDAD 81
GS+ + S + F +A+
Sbjct 918 PGSYYKINESMIKFHTENAE 937
> ath:AT1G68020 ATTPS6; ATTPS6; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=700
Score = 33.1 bits (74), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLT 31
R L DWF + +G+ AEHGY+ R T
Sbjct 650 RETLSDWFSPCEKLGIAAEHGYFLRYSTKT 679
> sce:YMR261C TPS3; Regulatory subunit of trehalose-6-phosphate
synthase/phosphatase complex, which synthesizes the storage
carbohydrate trehalose; expression is induced by stress conditions
and repressed by the Ras-cAMP pathway; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=1054
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 37/80 (46%), Gaps = 6/80 (7%)
Query 2 RSLLEDWFRDVKGIGLCAEHGYYYRLDKLTGDEWGCMCPDADFTWKSVAVELMLQYVKRT 61
++ E + V IGL AE+G Y R++ W + + D W ++ + V+R
Sbjct 829 KNTFESLYNGVLNIGLIAENGAYVRVN----GSWYNIVEELD--WMKEVAKIFDEKVERL 882
Query 62 QGSFTENKGSALVFQYRDAD 81
GS+ + S + F +AD
Sbjct 883 PGSYYKIADSMIRFHTENAD 902
> xla:734182 ara-55; Hic-5
Length=459
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 68 NKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGKAV 127
NKGS VF+ RD + D+ L ++ V+S G++EVK+ VN K
Sbjct 119 NKGSEEVFKPRDTEDPSSPRDTLDVPKALEDIPSPKSSEVMSTPGHMEVKIDQVNSDKVT 178
Query 128 ERIL 131
L
Sbjct 179 ASRL 182
> tgo:TGME49_093550 hypothetical protein
Length=899
Score = 29.6 bits (65), Expect = 4.4, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 34/73 (46%), Gaps = 11/73 (15%)
Query 86 QLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGKAVERILQTLSTLHGDIDFVF 145
Q + L H+ ++ Y V+ +GY E +LLG V+ + + + LH ++D
Sbjct 120 QQTERRLQRHIDDICLRYDSMVMEAQGYAESRLLG-----QVKDLQRVIWILHSELD--- 171
Query 146 CIGDDRSDEDMFA 158
+ R D+FA
Sbjct 172 ---ELRFQRDIFA 181
Lambda K H
0.321 0.140 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3832864436
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40