bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1795_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
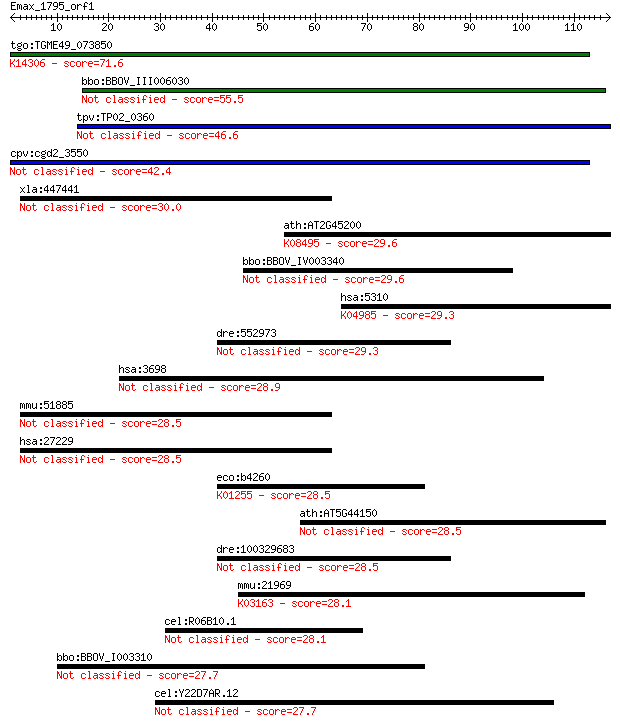
tgo:TGME49_073850 hypothetical protein ; K14306 nuclear pore c... 71.6 6e-13
bbo:BBOV_III006030 17.m07532; hypothetical protein 55.5 4e-08
tpv:TP02_0360 sporozoite and macroschizont protein 1 46.6
cpv:cgd2_3550 hypothetical protein 42.4 4e-04
xla:447441 tubgcp4, 76p, MGC81229; tubulin, gamma complex asso... 30.0 1.7
ath:AT2G45200 GOS12; GOS12 (GOLGI SNARE 12); SNARE binding; K0... 29.6 2.5
bbo:BBOV_IV003340 21.m03111; hypothetical protein 29.6 2.6
hsa:5310 PKD1, PBP, Pc-1, TRPP1; polycystic kidney disease 1 (... 29.3 2.8
dre:552973 npat, im:6901964; nuclear protein, ataxia-telangiec... 29.3 3.4
hsa:3698 ITIH2, H2P, SHAP; inter-alpha (globulin) inhibitor H2 28.9 4.3
mmu:51885 Tubgcp4, 4932441P04Rik, 76p, D2Ertd435e, MGC28085; t... 28.5 5.1
hsa:27229 TUBGCP4, 76P, FLJ14797, GCP4; tubulin, gamma complex... 28.5 5.1
eco:b4260 pepA, carP, ECK4253, JW4217, xerB; multifunctional a... 28.5 5.3
ath:AT5G44150 hypothetical protein 28.5 5.3
dre:100329683 Npat protein-like 28.5 5.4
mmu:21969 Top1, AI467334, D130064I21Rik, Top-1; topoisomerase ... 28.1 6.3
cel:R06B10.1 hypothetical protein 28.1 7.4
bbo:BBOV_I003310 19.m02357; ankyrin repeat domain containing p... 27.7 9.0
cel:Y22D7AR.12 hypothetical protein 27.7 9.9
> tgo:TGME49_073850 hypothetical protein ; K14306 nuclear pore
complex protein Nup62
Length=723
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 81/136 (59%), Gaps = 25/136 (18%)
Query 1 QQRVHRRQLFVGETIDMIEQQQNDISNLLASIEGSLLAKLSPQE---------------- 44
Q ++ +RQ ++ + ID +E+QQ D+ LLAS+E S+L ++ PQ+
Sbjct 573 QIKIEKRQTYICDFIDGLERQQRDLLTLLASVEASVLRQI-PQDNGDPTGAAGGDALAQR 631
Query 45 -QRSI-------CSTVEQSMSERVLDIDAQMDELSDAIAQAARRTQPDPVAAISQVLAVH 96
QR S+ E+ +S R+ +ID Q++++ A+++A R QP P+ ++QVL +H
Sbjct 632 VQREWEAESGFHSSSEEELLSRRLRNIDEQLNDVGLALSEATERFQPGPLGTVAQVLGIH 691
Query 97 QAALESATQQCAKLEQ 112
QAAL+++ +Q ++L+Q
Sbjct 692 QAALQASWRQASELQQ 707
> bbo:BBOV_III006030 17.m07532; hypothetical protein
Length=402
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 53/101 (52%), Gaps = 0/101 (0%)
Query 15 IDMIEQQQNDISNLLASIEGSLLAKLSPQEQRSICSTVEQSMSERVLDIDAQMDELSDAI 74
I +E++Q N L S+E SL +KL + RS Q++++++ ++ Q
Sbjct 294 IKHLEEEQLSAINTLDSMERSLKSKLEGRRGRSTSYQTVQNITKQLQNLQEQFSHAYKDA 353
Query 75 AQAARRTQPDPVAAISQVLAVHQAALESATQQCAKLEQCLK 115
A QP+P+ +++VL H A+L S QC +LE C+K
Sbjct 354 ENTAAVCQPEPLYTVAKVLTYHDASLSSLETQCLELENCIK 394
> tpv:TP02_0360 sporozoite and macroschizont protein 1
Length=900
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/103 (24%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 14 TIDMIEQQQNDISNLLASIEGSLLAKLSPQEQRSICSTVEQSMSERVLDIDAQMDELSDA 73
+I +E++Q ++L ++E L KL RS + Q+++ + ++ Q+ +
Sbjct 791 SIKTMEEEQKMALSVLDNMERVLKGKLETLNNRSNGYNMVQAITRNLQELSDQLSTTTKV 850
Query 74 IAQAARRTQPDPVAAISQVLAVHQAALESATQQCAKLEQCLKS 116
A QP+P+ I++VL+ HQ +L +QC ++++ +K+
Sbjct 851 AEDTAEACQPEPLYTIAKVLSFHQVSLIDLEKQCDEIDRRIKA 893
> cpv:cgd2_3550 hypothetical protein
Length=860
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 36/165 (21%), Positives = 58/165 (35%), Gaps = 53/165 (32%)
Query 1 QQRVHRRQLFVGETIDMIEQQQNDISNLLASIEGSLLAKLS------------------- 41
QQ + RQ + ID I QQQN +SN L IE S KL
Sbjct 684 QQEIRMRQENMDNRIDQIAQQQNSLSNTLKIIEESFSKKLEGSSSINSFSNNNNNNNNSN 743
Query 42 ----------------------------------PQEQRSICSTVEQSMSERVLDIDAQM 67
P + I + ++S + DI++Q+
Sbjct 744 NNNNNNSNNINNSNTNNIPRIGTTGGSFSGSHDKPNQNMRISAARANALSSELQDIESQV 803
Query 68 DELSDAIAQAARRTQPDPVAAISQVLAVHQAALESATQQCAKLEQ 112
+ L + R P P+ I ++L +HQ L+S + +L++
Sbjct 804 ETLMSQLNNLHERIYPSPLNEIVKILNMHQLTLQSIESEAQRLKE 848
> xla:447441 tubgcp4, 76p, MGC81229; tubulin, gamma complex associated
protein 4
Length=666
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 36/63 (57%), Gaps = 6/63 (9%)
Query 3 RVHRRQLFVGETIDMIEQQQNDIS---NLLASIEGSLLAKLSPQEQRSICSTVEQSMSER 59
RV + LFVGE++ M E Q ++S ++L + E + A+L +Q+ + S V+ E
Sbjct 271 RVAEKILFVGESVQMFENQNVNMSRTGSILKNQEDTFAAELHRLKQQPLFSLVD---FES 327
Query 60 VLD 62
VLD
Sbjct 328 VLD 330
> ath:AT2G45200 GOS12; GOS12 (GOLGI SNARE 12); SNARE binding;
K08495 golgi SNAP receptor complex member 1
Length=239
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 34/63 (53%), Gaps = 9/63 (14%)
Query 54 QSMSERVLDIDAQMDELSDAIAQAARRTQPDPVAAISQVLAVHQAALESATQQCAKLEQC 113
QS+ E++LDI+ D +S A AA P +++Q LA H+ L TQ+ +++
Sbjct 68 QSLLEKLLDIN---DSMSRCAASAA------PTTSVTQKLARHRDILHEYTQEFRRIKGN 118
Query 114 LKS 116
+ S
Sbjct 119 INS 121
> bbo:BBOV_IV003340 21.m03111; hypothetical protein
Length=433
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 25/52 (48%), Gaps = 1/52 (1%)
Query 46 RSICSTVEQSMSERVLDIDAQMDELSDAIAQAARRTQPDPVAAISQVLAVHQ 97
R + T+E + R+ D MD + D +A PD V ISQV A ++
Sbjct 231 RVLIKTLEAFCARRLKD-TGLMDTIGDVVASTLETLHPDEVVRISQVYARYR 281
> hsa:5310 PKD1, PBP, Pc-1, TRPP1; polycystic kidney disease 1
(autosomal dominant); K04985 polycystin 1
Length=4302
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query 65 AQMDELSDAIAQAARRTQPDP--VAAISQVLAVHQAALESATQQCAKLEQCLKS 116
+Q+D LS ++ + R +P+P + A+ + L L AT+ +LEQ L S
Sbjct 4191 SQLDGLSVSLGRLGTRCEPEPSRLQAVFEALLTQFDRLNQATEDVYQLEQQLHS 4244
> dre:552973 npat, im:6901964; nuclear protein, ataxia-telangiectasia
locus
Length=1310
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 41 SPQEQRSICSTVEQSMS-ERVLDIDAQMDELSDAIAQAARRTQPDP 85
SPQ ++ CSTVEQ S + +L + ++ DAI +T+ DP
Sbjct 285 SPQTSKATCSTVEQEQSIDEILGLQGEIHMTDDAIQDILAQTESDP 330
> hsa:3698 ITIH2, H2P, SHAP; inter-alpha (globulin) inhibitor
H2
Length=946
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 3/84 (3%)
Query 22 QNDISNLLASIEGSLLAKLSPQEQRSICS--TVEQSMSERVLDIDAQMDELSDAIAQAAR 79
QN+ N E + K P + I S T + ++ VL+ AQMD+L D +++ +
Sbjct 515 QNNFHNYFGGSEIVVAGKFDPAKLDQIESVITATSANTQLVLETLAQMDDLQDFLSK-DK 573
Query 80 RTQPDPVAAISQVLAVHQAALESA 103
PD + L ++Q E +
Sbjct 574 HADPDFTRKLWAYLTINQLLAERS 597
> mmu:51885 Tubgcp4, 4932441P04Rik, 76p, D2Ertd435e, MGC28085;
tubulin, gamma complex associated protein 4
Length=667
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 37/63 (58%), Gaps = 6/63 (9%)
Query 3 RVHRRQLFVGETIDMIEQQQNDIS---NLLASIEGSLLAKLSPQEQRSICSTVEQSMSER 59
RV + LFVGE++ M E Q +++ ++L + E + A+L +Q+ + S V+ E+
Sbjct 271 RVAEKILFVGESVQMFENQNVNLTRKGSILKNQEDTFAAELHRLKQQPLFSLVD---FEQ 327
Query 60 VLD 62
V+D
Sbjct 328 VVD 330
> hsa:27229 TUBGCP4, 76P, FLJ14797, GCP4; tubulin, gamma complex
associated protein 4
Length=666
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 37/63 (58%), Gaps = 6/63 (9%)
Query 3 RVHRRQLFVGETIDMIEQQQNDIS---NLLASIEGSLLAKLSPQEQRSICSTVEQSMSER 59
RV + LFVGE++ M E Q +++ ++L + E + A+L +Q+ + S V+ E+
Sbjct 271 RVAEKILFVGESVQMFENQNVNLTRKGSILKNQEDTFAAELHRLKQQPLFSLVD---FEQ 327
Query 60 VLD 62
V+D
Sbjct 328 VVD 330
> eco:b4260 pepA, carP, ECK4253, JW4217, xerB; multifunctional
aminopeptidase A: a cyteinylglycinase, transcription regulator
and site-specific recombination factor (EC:3.4.11.1); K01255
leucyl aminopeptidase [EC:3.4.11.1]
Length=503
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 41 SPQEQRSICSTVEQSMSERVLDIDAQMDELSDAIAQAARR 80
SP++QRS C V R+ I Q+D++SD A R
Sbjct 9 SPEKQRSACIVVGVFEPRRLSPIAEQLDKISDGYISALLR 48
> ath:AT5G44150 hypothetical protein
Length=355
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 2/59 (3%)
Query 57 SERVLDIDAQMDELSDAIAQAARRTQPDPVAAISQVLAVHQAALESATQQCAKLEQCLK 115
SE+ I+A +D L ++ ++A TQP+PVA+ S + + L+S + + EQ K
Sbjct 248 SEKSSAIEADLDLLLNSFSEA--HTQPNPVASASGKSSAFETELDSLLKSHSSTEQFNK 304
> dre:100329683 Npat protein-like
Length=1310
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 41 SPQEQRSICSTVEQSMS-ERVLDIDAQMDELSDAIAQAARRTQPDP 85
SPQ ++ CSTVEQ S + +L + ++ DAI +T+ DP
Sbjct 285 SPQTSKAACSTVEQEQSIDEILGLQGEIHMTDDAIQDILAQTESDP 330
> mmu:21969 Top1, AI467334, D130064I21Rik, Top-1; topoisomerase
(DNA) I (EC:5.99.1.2); K03163 DNA topoisomerase I [EC:5.99.1.2]
Length=767
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 36/72 (50%), Gaps = 7/72 (9%)
Query 45 QRSICSTVEQSMSERVLDIDAQMDELSDA-----IAQAARRTQPDPVAAISQVLAVHQAA 99
QR+ T E+SM IDA+ D+L+DA A+A + D A +V+ + A
Sbjct 635 QRAPPKTFEKSMMNLQSKIDAKKDQLADARRDLKSAKADAKVMKD--AKTKKVVESKKKA 692
Query 100 LESATQQCAKLE 111
++ +Q KLE
Sbjct 693 VQRLEEQLMKLE 704
> cel:R06B10.1 hypothetical protein
Length=1236
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 31 SIEGSLLAKLSPQEQRSICSTVEQSMSERVLDIDAQMD 68
S+ +L KL + +S+CSTV++ MS+ LD Q +
Sbjct 651 SMNEALNMKLDVNDLKSLCSTVQKEMSKVTLDAKDQQN 688
> bbo:BBOV_I003310 19.m02357; ankyrin repeat domain containing
protein
Length=726
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 10 FVGETIDMIEQQQNDISNLLASIEGSLLAKLSPQEQRSICSTVEQSMSERVLDIDAQMDE 69
++ ET+++ E+ +DI+ +LAS L + + + S + +S+ V D+ Q++
Sbjct 604 YLKETLNIFEEDPDDINRVLASTIAVALQEPNSDQIYRCISRIGPELSKTVFDLTMQIER 663
Query 70 LSDA-IAQAARR 80
IA +RR
Sbjct 664 NGGVLIADGSRR 675
> cel:Y22D7AR.12 hypothetical protein
Length=1137
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 20/89 (22%), Positives = 44/89 (49%), Gaps = 16/89 (17%)
Query 29 LASIEGSLLAKLSPQEQRSICSTVEQSMSERVLD------------IDAQMDELSDAIAQ 76
++S++ +L K+ + + +C+TV++ MS+ LD + A +D + ++ Q
Sbjct 567 ISSMDAALNMKIDLTKLKLLCATVQKEMSKVTLDAKDQKNLQRLATLGADLDRMKASLNQ 626
Query 77 AARRTQPDPVAAISQVLAVHQAALESATQ 105
+ +P + S LA H E+A++
Sbjct 627 SVSNVKP----SNSPKLADHAEVFEAASK 651
Lambda K H
0.314 0.124 0.322
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40