bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1774_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
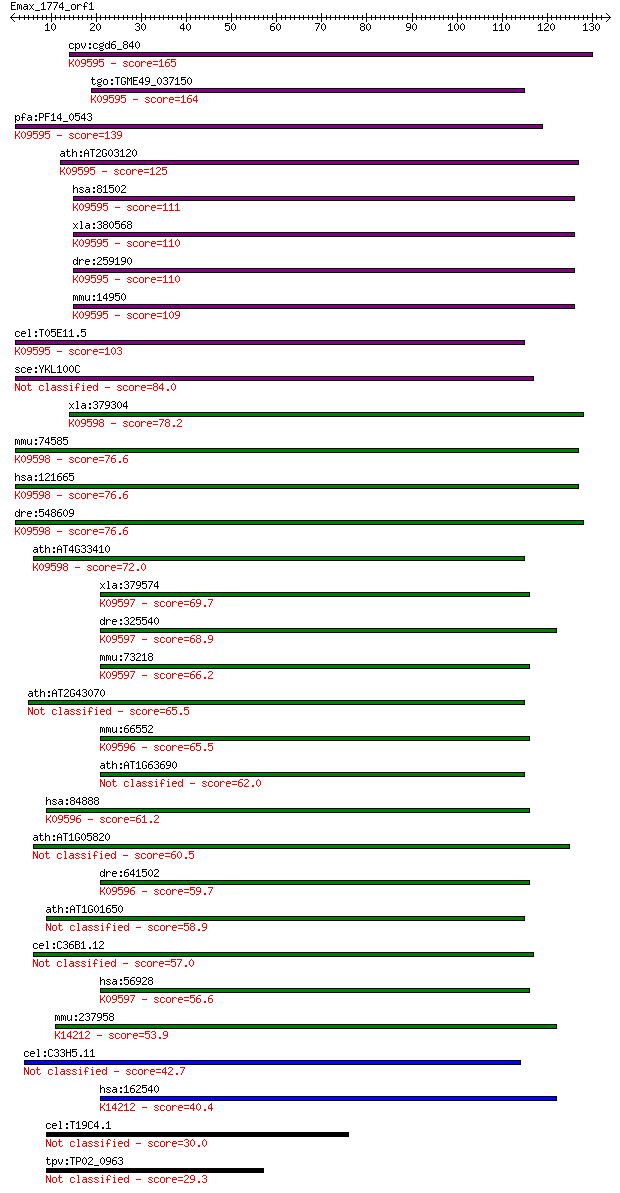
cpv:cgd6_840 shanti/Ykl100cp/Minor histocompatibility antigen ... 165 3e-41
tgo:TGME49_037150 signal peptide peptidase domain-containing p... 164 5e-41
pfa:PF14_0543 SPP; signal peptide peptidase; K09595 minor hist... 139 3e-33
ath:AT2G03120 ATSPP; ATSPP (ARABIDOPSIS SIGNAL PEPTIDE PEPTIDA... 125 2e-29
hsa:81502 HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3, PS... 111 8e-25
xla:380568 hm13, MGC68919, h13; histocompatibility (minor) 13 ... 110 1e-24
dre:259190 hm13, H13, SPP, cb228, wu:fc11a06, zgc:56660, zgc:8... 110 2e-24
mmu:14950 H13, 1200006O09Rik, 4930443L17Rik, 5031424B04Rik, AV... 109 2e-24
cel:T05E11.5 imp-2; IntraMembrane Protease (IMPAS) family memb... 103 1e-22
sce:YKL100C Putative protein of unknown function with similari... 84.0 1e-16
xla:379304 sppl3, MGC52975; signal peptide peptidase 3; K09598... 78.2 6e-15
mmu:74585 Sppl3, 4833416I09Rik, MGC90867, Psl4, Usmg3; signal ... 76.6 2e-14
hsa:121665 SPPL3, DKFZp586C1324, IMP2, MDHV1887, MGC126674, MG... 76.6 2e-14
dre:548609 sppl3, fb94d08, wu:fb94d08, zgc:114093; signal pept... 76.6 2e-14
ath:AT4G33410 signal peptide peptidase family protein; K09598 ... 72.0 5e-13
xla:379574 sppl2b, MGC69113; signal peptide peptidase-like 2B;... 69.7 2e-12
dre:325540 sppl2, SPPL2B, wu:fc16e01, wu:fc85d12, zgc:136525; ... 68.9 4e-12
mmu:73218 3110056O03Rik, AW550292, PSL1, Sppl2b; RIKEN cDNA 31... 66.2 3e-11
ath:AT2G43070 protease-associated (PA) domain-containing protein 65.5 4e-11
mmu:66552 2010106G01Rik, C130089K23Rik, Sppl2A; RIKEN cDNA 201... 65.5 4e-11
ath:AT1G63690 protease-associated (PA) domain-containing protein 62.0 5e-10
hsa:84888 SPPL2A, IMP3, PSL2; signal peptide peptidase-like 2A... 61.2 8e-10
ath:AT1G05820 aspartic-type endopeptidase/ peptidase 60.5 1e-09
dre:641502 MGC123258; zgc:123258; K09596 signal peptide peptid... 59.7 2e-09
ath:AT1G01650 aspartic-type endopeptidase/ peptidase 58.9 4e-09
cel:C36B1.12 imp-1; IntraMembrane Protease (IMPAS) family memb... 57.0 2e-08
hsa:56928 SPPL2B, IMP4, KIAA1532, MGC111084, PSL1; signal pept... 56.6 2e-08
mmu:237958 4933407P14Rik, IMP5, Sppl2c; RIKEN cDNA 4933407P14 ... 53.9 1e-07
cel:C33H5.11 imp-3; IntraMembrane Protease (IMPAS) family memb... 42.7 3e-04
hsa:162540 IMP5, SPPL2c; intramembrane protease 5; K14212 sign... 40.4 0.001
cel:T19C4.1 hypothetical protein 30.0 2.0
tpv:TP02_0963 hypothetical protein 29.3 3.0
> cpv:cgd6_840 shanti/Ykl100cp/Minor histocompatibility antigen
H13-like; presenilin, signal peptide peptidase family, with
10 transmembrane domains and a signal peptide ; K09595 minor
histocompatibility antigen H13 [EC:3.4.23.-]
Length=408
Score = 165 bits (418), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 72/116 (62%), Positives = 95/116 (81%), Gaps = 0/116 (0%)
Query 14 LWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAK 73
+W++T W +HN+ AIAFCIQAI+L+S+GSF + +ILLCGLFVYD+ WVFGT+VMV+VAK
Sbjct 187 IWIITDSWIIHNLFAIAFCIQAISLISIGSFKIGAILLCGLFVYDIFWVFGTDVMVTVAK 246
Query 74 AFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATAQQL 129
+F+GPAKL+FPV P + SILGLGD+VIPG+ I++CLRFDL Y K+ N + L
Sbjct 247 SFQGPAKLIFPVSFDPWKQSILGLGDIVIPGLFISLCLRFDLKDYTKKHNQSLYHL 302
> tgo:TGME49_037150 signal peptide peptidase domain-containing
protein ; K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=417
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 75/96 (78%), Positives = 88/96 (91%), Gaps = 0/96 (0%)
Query 19 KHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAKAFEGP 78
KHWALHN+L IAFCIQAI+LVSVG+F VA+ILL GLF+YD+ WVFGT+VMV+VAK+FEGP
Sbjct 209 KHWALHNLLGIAFCIQAISLVSVGNFTVATILLSGLFIYDIFWVFGTDVMVTVAKSFEGP 268
Query 79 AKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRFD 114
AKL+FPV I P Q+SILGLGD+VIPGV I+MCLRFD
Sbjct 269 AKLIFPVSIHPWQHSILGLGDIVIPGVFISMCLRFD 304
> pfa:PF14_0543 SPP; signal peptide peptidase; K09595 minor histocompatibility
antigen H13 [EC:3.4.23.-]
Length=412
Score = 139 bits (349), Expect = 3e-33, Method: Composition-based stats.
Identities = 61/117 (52%), Positives = 90/117 (76%), Gaps = 0/117 (0%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
I+ +++ AI W+ K + HN+LA++FC QAI+LV + +F + +LL GLFVYD+ W
Sbjct 172 IVCLILSFAIGLRWIFYKDFITHNVLAVSFCFQAISLVILSNFLIGFLLLSGLFVYDIFW 231
Query 62 VFGTEVMVSVAKAFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRFDLFLY 118
VFG +VMV+VAK+FE P KL+FPV P+ YS+LGLGD++IPG+L+++CLRFD +L+
Sbjct 232 VFGNDVMVTVAKSFEAPVKLLFPVSSDPVHYSMLGLGDIIIPGILMSLCLRFDYYLF 288
> ath:AT2G03120 ATSPP; ATSPP (ARABIDOPSIS SIGNAL PEPTIDE PEPTIDASE);
aspartic-type endopeptidase; K09595 minor histocompatibility
antigen H13 [EC:3.4.23.-]
Length=344
Score = 125 bits (315), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 59/115 (51%), Positives = 80/115 (69%), Gaps = 0/115 (0%)
Query 12 CTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSV 71
C + KHW +NIL ++FCIQ I ++S+GSF +ILL GLF YD+ WVF T VMVSV
Sbjct 152 CAWYAWKKHWLANNILGLSFCIQGIEMLSLGSFKTGAILLAGLFFYDIFWVFFTPVMVSV 211
Query 72 AKAFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATA 126
AK+F+ P KL+FP + YS+LGLGD+VIPG+ +A+ LRFD+ + Q T+
Sbjct 212 AKSFDAPIKLLFPTGDALRPYSMLGLGDIVIPGIFVALALRFDVSRRRQPQYFTS 266
> hsa:81502 HM13, H13, IMP1, IMPAS, IMPAS-1, MSTP086, PSENL3,
PSL3, SPP, dJ324O17.1; histocompatibility (minor) 13; K09595
minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=377
Score = 111 bits (277), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 55/116 (47%), Positives = 76/116 (65%), Gaps = 7/116 (6%)
Query 15 WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAKA 74
+LL KHW +N+ +AF + + L+ + + ILL GLF+YDV WVFGT VMV+VAK+
Sbjct 176 YLLRKHWIANNLFGLAFSLNGVELLHLNNVSTGCILLGGLFIYDVFWVFGTNVMVTVAKS 235
Query 75 FEGPAKLMFPVQ-----ISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNAT 125
FE P KL+FP + +++LGLGDVVIPG+ IA+ LRFD + LK+ T
Sbjct 236 FEAPIKLVFPQDLLEKGLEANNFAMLGLGDVVIPGIFIALLLRFD--ISLKKNTHT 289
> xla:380568 hm13, MGC68919, h13; histocompatibility (minor) 13
(EC:3.4.99.-); K09595 minor histocompatibility antigen H13
[EC:3.4.23.-]
Length=392
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 54/116 (46%), Positives = 77/116 (66%), Gaps = 7/116 (6%)
Query 15 WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAKA 74
+LL KHW +N+ +AF + + L+ + + ILL GLFVYD+ WVFGT VMV+VAK+
Sbjct 174 YLLKKHWIANNLFGLAFSLNGVELLHLNNVSTGCILLGGLFVYDIFWVFGTNVMVTVAKS 233
Query 75 FEGPAKLMFPVQ-----ISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNAT 125
FE P KL+FP + +++LGLGD+VIPG+ IA+ LRFD + LK+ + T
Sbjct 234 FEAPIKLVFPQDLLEKGLEANNFAMLGLGDIVIPGIFIALLLRFD--VSLKKNSHT 287
> dre:259190 hm13, H13, SPP, cb228, wu:fc11a06, zgc:56660, zgc:86862;
histocompatibility (minor) 13 (EC:3.4.99.-); K09595
minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=366
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/116 (46%), Positives = 76/116 (65%), Gaps = 7/116 (6%)
Query 15 WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAKA 74
++L KHW +N+ +AF + + L+ + + ILL GLFVYDV WVFGT VMV+VAK+
Sbjct 179 YVLKKHWIANNLFGLAFALNGVELLHLNNVSTGCILLGGLFVYDVFWVFGTNVMVTVAKS 238
Query 75 FEGPAKLMFPVQ-----ISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNAT 125
FE P KL+FP + +++LGLGD+VIPG+ IA+ LRFD + LK+ T
Sbjct 239 FEAPIKLVFPQDLLEKGLGASNFAMLGLGDIVIPGIFIALLLRFD--VSLKKNTRT 292
> mmu:14950 H13, 1200006O09Rik, 4930443L17Rik, 5031424B04Rik,
AV020344, H-13, Hm13, PSL3, Spp; histocompatibility 13 (EC:3.4.99.-);
K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=394
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 53/116 (45%), Positives = 76/116 (65%), Gaps = 7/116 (6%)
Query 15 WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVMVSVAKA 74
+LL KHW +N+ +AF + + L+ + + ILL GLF+YD+ WVFGT VMV+VAK+
Sbjct 176 YLLRKHWIANNLFGLAFSLNGVELLHLNNVSTGCILLGGLFIYDIFWVFGTNVMVTVAKS 235
Query 75 FEGPAKLMFPVQ-----ISPLQYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNAT 125
FE P KL+FP + +++LGLGD+VIPG+ IA+ LRFD + LK+ T
Sbjct 236 FEAPIKLVFPQDLLEKGLEADNFAMLGLGDIVIPGIFIALLLRFD--ISLKKNTHT 289
> cel:T05E11.5 imp-2; IntraMembrane Protease (IMPAS) family member
(imp-2); K09595 minor histocompatibility antigen H13 [EC:3.4.23.-]
Length=468
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 54/119 (45%), Positives = 73/119 (61%), Gaps = 6/119 (5%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
II+ L+ I LL +HW +NI+ ++F I I + + SF S+LL GLF YD+ W
Sbjct 250 IIALLMCSPILISHLLKRHWITNNIIGVSFSILGIERLHLASFKAGSLLLVGLFFYDIFW 309
Query 62 VFGTEVMVSVAKAFEGPAKLMFPVQI------SPLQYSILGLGDVVIPGVLIAMCLRFD 114
VFGT+VM SVAK + P L FP I ++S+LGLGD+VIPG+ IA+ RFD
Sbjct 310 VFGTDVMTSVAKGIDAPILLQFPQDIYRNGIMEASKHSMLGLGDIVIPGIFIALLRRFD 368
> sce:YKL100C Putative protein of unknown function with similarity
to a human minor histocompatibility antigen and signal
peptide peptidases; YKL100C is not an essential gene
Length=587
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 42/120 (35%), Positives = 76/120 (63%), Gaps = 5/120 (4%)
Query 2 IISHLVALAICTLWLLTKH-WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVI 60
I+S ++++ + L+ + W + N +++ I +IA + + + +++L LF YD+
Sbjct 309 IVSFVLSIVSTVYFYLSPNDWLISNAVSMNMAIWSIAQLKLKNLKSGALILIALFFYDIC 368
Query 61 WVFGTEVMVSVAKAFEGPAKLMFPVQISPLQ----YSILGLGDVVIPGVLIAMCLRFDLF 116
+VFGT+VMV+VA + P KL PV+ + Q +SILGLGD+ +PG+ IAMC ++D++
Sbjct 369 FVFGTDVMVTVATNLDIPVKLSLPVKFNTAQNNFNFSILGLGDIALPGMFIAMCYKYDIW 428
> xla:379304 sppl3, MGC52975; signal peptide peptidase 3; K09598
signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=379
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 50/145 (34%), Positives = 71/145 (48%), Gaps = 34/145 (23%)
Query 14 LWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWV------FGTEV 67
+W+LT HW L + LA+ C+ IA V + S V+ +LL GL +YDV WV F + V
Sbjct 155 VWVLTGHWLLMDALAMGLCVAMIAFVRLPSLKVSCLLLSGLLIYDVFWVFFSAYIFSSNV 214
Query 68 MVSVA-------------------------KAFEGPAKLMFPVQISPLQYSILGLGDVVI 102
MV VA P KL+FP +S+LG+GD+V+
Sbjct 215 MVKVATQPADNPLDVLSRKLHLGPNVGRDVPRLSLPGKLVFPSSTGS-HFSMLGIGDIVM 273
Query 103 PGVLIAMCLRFDLFLYLKQQNATAQ 127
PG+L+ LR+D Y KQ + +Q
Sbjct 274 PGLLLCFVLRYD--NYKKQATSDSQ 296
> mmu:74585 Sppl3, 4833416I09Rik, MGC90867, Psl4, Usmg3; signal
peptide peptidase 3; K09598 signal peptide peptidase-like
3 [EC:3.4.23.-]
Length=384
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/156 (32%), Positives = 77/156 (49%), Gaps = 34/156 (21%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
++S +++ + +W+LT HW L + LA+ C+ IA V + S V+ +LL GL +YDV W
Sbjct 144 LLSFSLSVMLVLIWVLTGHWLLMDALAMGLCVAMIAFVRLPSLKVSCLLLSGLLIYDVFW 203
Query 62 V------FGTEVMVSVA-------------------------KAFEGPAKLMFPVQISPL 90
V F + VMV VA P KL+FP
Sbjct 204 VFFSAYIFNSNVMVKVATQPADNPLDVLSRKLHLGPNVGRDVPRLSLPGKLVFPSSTGS- 262
Query 91 QYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATA 126
+S+LG+GD+V+PG+L+ LR+D Y KQ + +
Sbjct 263 HFSMLGIGDIVMPGLLLCFVLRYD--NYKKQASGDS 296
> hsa:121665 SPPL3, DKFZp586C1324, IMP2, MDHV1887, MGC126674,
MGC126676, MGC90402, PRO4332, PSL4; signal peptide peptidase
3; K09598 signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=384
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/156 (32%), Positives = 77/156 (49%), Gaps = 34/156 (21%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
++S +++ + +W+LT HW L + LA+ C+ IA V + S V+ +LL GL +YDV W
Sbjct 144 LLSFSLSVMLVLIWVLTGHWLLMDALAMGLCVAMIAFVRLPSLKVSCLLLSGLLIYDVFW 203
Query 62 V------FGTEVMVSVA-------------------------KAFEGPAKLMFPVQISPL 90
V F + VMV VA P KL+FP
Sbjct 204 VFFSAYIFNSNVMVKVATQPADNPLDVLSRKLHLGPNVGRDVPRLSLPGKLVFPSSTGS- 262
Query 91 QYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATA 126
+S+LG+GD+V+PG+L+ LR+D Y KQ + +
Sbjct 263 HFSMLGIGDIVMPGLLLCFVLRYD--NYKKQASGDS 296
> dre:548609 sppl3, fb94d08, wu:fb94d08, zgc:114093; signal peptide
peptidase 3; K09598 signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=382
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 49/157 (31%), Positives = 78/157 (49%), Gaps = 36/157 (22%)
Query 2 IISHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
++S +++ + +W+LT HW L + LA+ C+ IA V + S V+ +LL GL +YDV W
Sbjct 145 LLSFSLSVMLVLIWVLTGHWLLMDALAMGLCVAMIAFVRLPSLKVSCLLLSGLLIYDVFW 204
Query 62 V------FGTEVMVSVA-------------------------KAFEGPAKLMFPVQISPL 90
V F + VMV VA P KL+FP
Sbjct 205 VFFSAYIFNSNVMVKVATQPADNPLDVLSRKLHLGPGMGRDVPRLSLPGKLVFPSSTGS- 263
Query 91 QYSILGLGDVVIPGVLIAMCLRFDLFLYLKQQNATAQ 127
+S+LG+GD+V+PG+L+ LR+D + ++ AT +
Sbjct 264 HFSMLGIGDIVMPGLLLCFVLRYDNY----KKQATGE 296
> ath:AT4G33410 signal peptide peptidase family protein; K09598
signal peptide peptidase-like 3 [EC:3.4.23.-]
Length=372
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 50/148 (33%), Positives = 75/148 (50%), Gaps = 39/148 (26%)
Query 6 LVALAICTL-WLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWV-- 62
LVA A+ + WL++ HW L+N+L I+ CI ++ V + + + ++LL LFVYD+ WV
Sbjct 133 LVACAMTVVAWLISGHWVLNNLLGISICIAFVSHVRLPNIKICAMLLVCLFVYDIFWVFF 192
Query 63 ----FGTEVMVSVA------------------------KAFEGPAKLMFPVQ-------- 86
FG VMV+VA K E P K++FP
Sbjct 193 SERFFGANVMVAVATQQASNPVHTVANSLNLPGLQLITKKLELPVKIVFPRNLLGGVVPG 252
Query 87 ISPLQYSILGLGDVVIPGVLIAMCLRFD 114
+S + +LGLGD+ IP +L+A+ L FD
Sbjct 253 VSASDFMMLGLGDMAIPAMLLALVLCFD 280
> xla:379574 sppl2b, MGC69113; signal peptide peptidase-like 2B;
K09597 signal peptide peptidase-like 2B [EC:3.4.23.-]
Length=606
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 43/116 (37%), Positives = 62/116 (53%), Gaps = 21/116 (18%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L +IL IAFC+ + + + +F ++LL LFVYDV +VF G +MV VA
Sbjct 295 WVLQDILGIAFCLYMLKTIRMPTFKGCTLLLFVLFVYDVFFVFITPYLTKRGESIMVEVA 354
Query 73 KAFEG-------PAKLMFP-VQISPLQ-----YSILGLGDVVIPGVLIAMCLRFDL 115
P L P + SPL +S+LG GD+++PG+L+A C RFD+
Sbjct 355 SGPSNSTTQEKLPMVLKVPRLNSSPLALCDRPFSLLGFGDILVPGLLVAYCHRFDI 410
> dre:325540 sppl2, SPPL2B, wu:fc16e01, wu:fc85d12, zgc:136525;
signal peptide peptidase-like 2; K09597 signal peptide peptidase-like
2B [EC:3.4.23.-]
Length=564
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 43/122 (35%), Positives = 63/122 (51%), Gaps = 21/122 (17%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L + L IAFC+ + + + +F ++LL LFVYDV +VF G +MV VA
Sbjct 324 WILQDALGIAFCLYMLKTIRLPTFKACTLLLVVLFVYDVFFVFITPLLTKSGESIMVEVA 383
Query 73 KAFEG-------PAKLMFP-VQISPL-----QYSILGLGDVVIPGVLIAMCLRFDLFLYL 119
P L P + SPL +S+LG GD+++PG+L+A C RFD+ +
Sbjct 384 AGPSDSSTHEKLPMVLKVPRLNSSPLVLCDRPFSLLGFGDILVPGLLVAYCHRFDILMQT 443
Query 120 KQ 121
Q
Sbjct 444 SQ 445
> mmu:73218 3110056O03Rik, AW550292, PSL1, Sppl2b; RIKEN cDNA
3110056O03 gene (EC:3.4.23.-); K09597 signal peptide peptidase-like
2B [EC:3.4.23.-]
Length=578
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 61/116 (52%), Gaps = 21/116 (18%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L + L IAFC+ + + + +F ++LL LF+YD+ +VF G +MV VA
Sbjct 315 WVLQDTLGIAFCLYMLKTIRLPTFKACTLLLLVLFIYDIFFVFITPFLTKSGNSIMVEVA 374
Query 73 KAFEG-------PAKLMFP-VQISPLQ-----YSILGLGDVVIPGVLIAMCLRFDL 115
P L P + SPL +S+LG GD+++PG+L+A C RFD+
Sbjct 375 TGPSNSSTHEKLPMVLKVPRLNTSPLSLCDRPFSLLGFGDILVPGLLVAYCHRFDI 430
> ath:AT2G43070 protease-associated (PA) domain-containing protein
Length=540
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 63/126 (50%), Gaps = 16/126 (12%)
Query 5 HLVALAICTLWLLTKH----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVI 60
++V LA W + +H W +IL I I A+ +V + + VA++LLC FVYD+
Sbjct 330 NIVCLAFAVFWFIKRHTSYSWVGQDILGICLMITALQVVRLPNIKVATVLLCCAFVYDIF 389
Query 61 WV------FGTEVMVSVAKAFEG-----PAKLMFPVQISPL-QYSILGLGDVVIPGVLIA 108
WV F VM+ VA+ P L P P Y ++G GD++ PG+LI+
Sbjct 390 WVFISPLIFHESVMIVVAQGDSSTGESIPMLLRIPRFFDPWGGYDMIGFGDILFPGLLIS 449
Query 109 MCLRFD 114
R+D
Sbjct 450 FASRYD 455
> mmu:66552 2010106G01Rik, C130089K23Rik, Sppl2A; RIKEN cDNA 2010106G01
gene (EC:3.4.23.-); K09596 signal peptide peptidase-like
2A [EC:3.4.23.-]
Length=523
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 42/115 (36%), Positives = 60/115 (52%), Gaps = 20/115 (17%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L +IL IAFC+ I + + +F ILL L +YDV +VF G +MV +A
Sbjct 318 WILQDILGIAFCLNLIKTMKLPNFMSCVILLGLLLIYDVFFVFITPFITKNGESIMVELA 377
Query 73 KA-FEGPAKLMFPVQISPLQ-----------YSILGLGDVVIPGVLIAMCLRFDL 115
FE KL +++ L S+LG GD+++PG+LIA C RFD+
Sbjct 378 AGPFENAEKLPVVIRVPKLMGYSVMSVCSVPVSVLGFGDIIVPGLLIAYCRRFDV 432
> ath:AT1G63690 protease-associated (PA) domain-containing protein
Length=540
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 58/106 (54%), Gaps = 12/106 (11%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTE------VMVSVAKA 74
W ++L IA I + +V V + V ++LL F+YD+ WVF ++ VM+ VA+
Sbjct 348 WIGQDVLGIALIITVLQIVHVPNLKVGTVLLSCAFLYDIFWVFVSKKLFHESVMIVVARG 407
Query 75 FEG-----PAKLMFPVQISPL-QYSILGLGDVVIPGVLIAMCLRFD 114
+ P L P P YSI+G GD+++PG+LIA LR+D
Sbjct 408 DKSGEDGIPMLLKIPRMFDPWGGYSIIGFGDILLPGLLIAFALRYD 453
> hsa:84888 SPPL2A, IMP3, PSL2; signal peptide peptidase-like
2A (EC:3.4.23.-); K09596 signal peptide peptidase-like 2A [EC:3.4.23.-]
Length=520
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 68/132 (51%), Gaps = 25/132 (18%)
Query 9 LAICTLWLLTKH-----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF 63
+A+ +W + ++ W L +IL IAFC+ I + + +F ILL L +YDV +VF
Sbjct 297 IAVAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFKSCVILLGLLLLYDVFFVF 356
Query 64 --------GTEVMVSVAKA-FEGPAKLMFPVQISPLQY-----------SILGLGDVVIP 103
G +MV +A F KL +++ L Y SILG GD+++P
Sbjct 357 ITPFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVP 416
Query 104 GVLIAMCLRFDL 115
G+LIA C RFD+
Sbjct 417 GLLIAYCRRFDV 428
> ath:AT1G05820 aspartic-type endopeptidase/ peptidase
Length=507
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 62/135 (45%), Gaps = 21/135 (15%)
Query 6 LVALAICTLWLLTKH----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
L + LW + + WA +I I I + + + + VA+ILLC F YD+ W
Sbjct 328 LFCFVVAILWFMNRKTSHAWAGQDIFGICMMINVLQVARLPNIRVATILLCCAFFYDIFW 387
Query 62 V------FGTEVMVSVAKAFEG-----PAKLMFPVQISPL-QYSILGLGDVVIPGVLIAM 109
V F VM++VA+ + P L P P Y+++G GD++ PG+LI
Sbjct 388 VFISPLIFKQSVMIAVARGSKDTGESIPMLLRIPRLSDPWGGYNMIGFGDILFPGLLICF 447
Query 110 CLRFDLFLYLKQQNA 124
RFD K+ N
Sbjct 448 IFRFD-----KENNK 457
> dre:641502 MGC123258; zgc:123258; K09596 signal peptide peptidase-like
2A [EC:3.4.23.-]
Length=519
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 46/132 (34%), Positives = 65/132 (49%), Gaps = 37/132 (28%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L ++L IAFC+ + +S+ +F + ILL L +YDV +VF G +MV VA
Sbjct 310 WVLQDLLGIAFCLNFLKTISLSNFKICVILLSLLLLYDVFFVFITPFLTPNGESIMVQVA 369
Query 73 ------------KAFEGPAK----------LMFPVQISPL-------QYSILGLGDVVIP 103
+ E PA +M Q S L Q+SILG GD++IP
Sbjct 370 LGPGGGGGKGDGRTVEVPADPSETYEKLPVVMRIPQFSALAQNLCMMQFSILGYGDIIIP 429
Query 104 GVLIAMCLRFDL 115
G+L+A C RFD+
Sbjct 430 GLLVAYCHRFDV 441
> ath:AT1G01650 aspartic-type endopeptidase/ peptidase
Length=398
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 60/122 (49%), Gaps = 16/122 (13%)
Query 9 LAICTLWLLTKH----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFG 64
+A W + + W +IL I+ I + +V V + V +LL F+YD+ WVF
Sbjct 192 IAFAVFWAVKRQYSYAWIGQDILGISLIITVLQIVRVPNLKVGFVLLSCAFMYDIFWVFV 251
Query 65 TE------VMVSVAKAFEG-----PAKLMFPVQISPLQ-YSILGLGDVVIPGVLIAMCLR 112
++ VM+ VA+ P L P P YSI+G GD+++PG+L+ LR
Sbjct 252 SKWWFRESVMIVVARGDRSGEDGIPMLLKIPRMFDPWGGYSIIGFGDIILPGLLVTFALR 311
Query 113 FD 114
+D
Sbjct 312 YD 313
> cel:C36B1.12 imp-1; IntraMembrane Protease (IMPAS) family member
(imp-1)
Length=662
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 41/157 (26%), Positives = 66/157 (42%), Gaps = 46/157 (29%)
Query 6 LVALAICTLWLLTKH----WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIW 61
++ L+ C W + + + L +++ +A C+ + + + S SIL+ +FVYD
Sbjct 377 IICLSFCVTWFIIRRQPYAFILLDVINMALCMHVLKCLRLPSLKWISILMLCMFVYDAFM 436
Query 62 VFGT--------EVMVSVAKAFEGPAK----------------------LMFPVQISPL- 90
VFGT VM+ VA AK LM +P+
Sbjct 437 VFGTPYMTTNGCSVMLEVATGLSCAAKGKNKGYPVPPIEQESVPEKFPMLMQVAHFNPMN 496
Query 91 -----------QYSILGLGDVVIPGVLIAMCLRFDLF 116
Q++ILGLGD+V+PG L+A C + F
Sbjct 497 ECLDMEIELGFQFTILGLGDIVMPGYLVAHCFTMNGF 533
> hsa:56928 SPPL2B, IMP4, KIAA1532, MGC111084, PSL1; signal peptide
peptidase-like 2B (EC:3.4.23.-); K09597 signal peptide
peptidase-like 2B [EC:3.4.23.-]
Length=511
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 62/116 (53%), Gaps = 21/116 (18%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L + L IAFC+ + + + +F ++LL LF+YD+ +VF G+ +MV VA
Sbjct 322 WVLQDALGIAFCLYMLKTIRLPTFKACTLLLLVLFLYDIFFVFITPFLTKSGSSIMVEVA 381
Query 73 KAFEG-------PAKLMFP-VQISPL-----QYSILGLGDVVIPGVLIAMCLRFDL 115
P L P + SPL +S+LG GD+++PG+L+A C RFD+
Sbjct 382 TGPSDSATREKLPMVLKVPRLNSSPLALCDRPFSLLGFGDILVPGLLVAYCHRFDI 437
> mmu:237958 4933407P14Rik, IMP5, Sppl2c; RIKEN cDNA 4933407P14
gene; K14212 signal peptide peptidase-like 2C [EC:3.4.23.-]
Length=581
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 43/140 (30%), Positives = 70/140 (50%), Gaps = 32/140 (22%)
Query 11 ICTLWLLTK---HWA--LHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF-- 63
+ LW++ + HWA L + L +A+C+ + V + +F ++ L L +DV +VF
Sbjct 343 VTVLWVIHRNEDHWAWLLQDTLGVAYCLFVLRRVRLPTFKNCTLFLLALLAFDVFFVFIT 402
Query 64 ------GTEVMVSVAKAFEGPA------KLMFPVQISPLQYS----------ILGLGDVV 101
G +MV VA GPA +L +++ L +S ILG GD+V
Sbjct 403 PLFTKTGESIMVEVAS---GPADSSSHERLPMVLKVPRLSFSALTLCNQPFSILGFGDIV 459
Query 102 IPGVLIAMCLRFDLFLYLKQ 121
+PG L+A C RFD+ + +Q
Sbjct 460 VPGFLVAYCHRFDMQVQSRQ 479
> cel:C33H5.11 imp-3; IntraMembrane Protease (IMPAS) family member
(imp-3)
Length=516
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 25/110 (22%), Positives = 57/110 (51%), Gaps = 1/110 (0%)
Query 4 SHLVALAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF 63
S LV+L+ + +W ++ILA A + + S+ A I + G+ ++D+ +++
Sbjct 343 SLLVSLSFGLYHECSGNWISNDILAFASIYVVCSRIQAVSYQTAIIFVIGMSLFDLFFLY 402
Query 64 GTEVMVSVAKAFEGPAKLMFPVQISPLQYSILGLGDVVIPGVLIAMCLRF 113
+++ +V K P ++ P + S+ L D+++PGV + + L++
Sbjct 403 VVDLLSTVVKENRSPLMILVPRDTKGNKQSLAAL-DIMVPGVFLNVVLKY 451
> hsa:162540 IMP5, SPPL2c; intramembrane protease 5; K14212 signal
peptide peptidase-like 2C [EC:3.4.23.-]
Length=684
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 40/125 (32%), Positives = 61/125 (48%), Gaps = 27/125 (21%)
Query 21 WALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVF--------GTEVMVSVA 72
W L + L I++C+ + V + + S L L +DV +VF G +M VA
Sbjct 349 WLLQDTLGISYCLFVLHRVRLPTLKNCSSFLLALLAFDVFFVFVTPFFTKTGESIMAQVA 408
Query 73 KAFEGPAK----------LMFP-VQISPL-----QYSILGLGDVVIPGVLIAMCLRFDLF 116
GPA+ L P +++S L +SILG GD+V+PG L+A C RFD+
Sbjct 409 L---GPAESSSHERLPMVLKVPRLRVSALTLCSQPFSILGFGDIVVPGFLVAYCCRFDVQ 465
Query 117 LYLKQ 121
+ +Q
Sbjct 466 VCSRQ 470
> cel:T19C4.1 hypothetical protein
Length=424
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 33/67 (49%), Gaps = 12/67 (17%)
Query 9 LAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFVYDVIWVFGTEVM 68
LAI T +L +W N+++I C+Q +A + + GV+ I Y +W G E++
Sbjct 248 LAIRTPYL---NW---NMISIVCCLQVMACLGIFLVGVSRIF------YGALWAIGIEII 295
Query 69 VSVAKAF 75
+ F
Sbjct 296 FATVALF 302
> tpv:TP02_0963 hypothetical protein
Length=810
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 9 LAICTLWLLTKHWALHNILAIAFCIQAIALVSVGSFGVASILLCGLFV 56
L IC WL +H L + C + L SV ++G + LL G+ V
Sbjct 510 LPICDQWLPLEHLNLEEWMLPQMCQHYLNLFSVSTYGSLAGLLVGIDV 557
Lambda K H
0.333 0.143 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40