bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1681_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
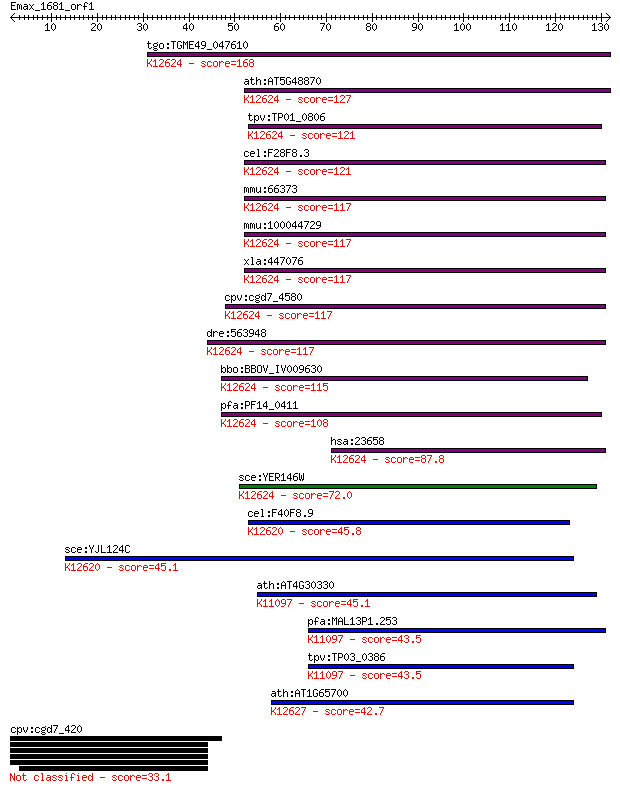
tgo:TGME49_047610 snRNP protein Lsm5, putative ; K12624 U6 snR... 168 4e-42
ath:AT5G48870 SAD1; SAD1 (SUPERSENSITIVE TO ABA AND DROUGHT 1)... 127 1e-29
tpv:TP01_0806 U6 small nuclear ribonucleoprotein E; K12624 U6 ... 121 5e-28
cel:F28F8.3 lsm-5; LSM Sm-like protein family member (lsm-5); ... 121 6e-28
mmu:66373 Lsm5, 2010208O10Rik, 2310034K10Rik; LSM5 homolog, U6... 117 7e-27
mmu:100044729 u6 snRNA-associated Sm-like protein LSm5-like; K... 117 7e-27
xla:447076 lsm5; LSM5 homolog, U6 small nuclear RNA associated... 117 7e-27
cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM dom... 117 9e-27
dre:563948 zgc:171959; K12624 U6 snRNA-associated Sm-like prot... 117 1e-26
bbo:BBOV_IV009630 23.m05873; snRNP Sm-like protein; K12624 U6 ... 115 4e-26
pfa:PF14_0411 small nuclear ribonucleoprotein, putative; K1262... 108 5e-24
hsa:23658 LSM5, FLJ12710, YER146W; LSM5 homolog, U6 small nucl... 87.8 7e-18
sce:YER146W LSM5; Lsm (Like Sm) protein; part of heteroheptame... 72.0 4e-13
cel:F40F8.9 lsm-1; LSM Sm-like protein family member (lsm-1); ... 45.8 4e-05
sce:YJL124C LSM1, SPB8; Lsm (Like Sm) protein; forms heterohep... 45.1 5e-05
ath:AT4G30330 small nuclear ribonucleoprotein E, putative / sn... 45.1 6e-05
pfa:MAL13P1.253 small nuclear ribonucleoprotein, putative; K11... 43.5 2e-04
tpv:TP03_0386 small nuclear ribonucleoprotein; K11097 small nu... 43.5 2e-04
ath:AT1G65700 small nuclear ribonucleoprotein, putative / snRN... 42.7 3e-04
cpv:cgd7_420 protein with DEXDc plus ring plus HELICc; possibl... 33.1 0.22
> tgo:TGME49_047610 snRNP protein Lsm5, putative ; K12624 U6 snRNA-associated
Sm-like protein LSm5
Length=119
Score = 168 bits (425), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 71/101 (70%), Positives = 91/101 (90%), Gaps = 0/101 (0%)
Query 31 NSSSSSSSSSNTAAGVLSGGPSYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNM 90
++++ ++S+ +GV+SGGPSYLPLALVDKC+GSR+W+IMKGDKEL GTLRGFDD+VNM
Sbjct 2 SAAAPKATSAPATSGVVSGGPSYLPLALVDKCIGSRMWIIMKGDKELAGTLRGFDDFVNM 61
Query 91 VLDDVTEYTFTAEGIKKNKLDTILLNGNNVSILVPGGNPEE 131
VLDDVTEYTFT G+KK KL +ILLNGN++++LVPGG+PEE
Sbjct 62 VLDDVTEYTFTPTGVKKTKLQSILLNGNSITMLVPGGDPEE 102
> ath:AT5G48870 SAD1; SAD1 (SUPERSENSITIVE TO ABA AND DROUGHT
1); RNA binding; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=88
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 56/80 (70%), Positives = 68/80 (85%), Gaps = 0/80 (0%)
Query 52 SYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLD 111
LP L+D+C+GS++WVIMKGDKELVG L+GFD YVNMVL+DVTEY TAEG + KLD
Sbjct 7 QLLPSELIDRCIGSKIWVIMKGDKELVGILKGFDVYVNMVLEDVTEYEITAEGRRVTKLD 66
Query 112 TILLNGNNVSILVPGGNPEE 131
ILLNGNN++ILVPGG+PE+
Sbjct 67 QILLNGNNIAILVPGGSPED 86
> tpv:TP01_0806 U6 small nuclear ribonucleoprotein E; K12624 U6
snRNA-associated Sm-like protein LSm5
Length=90
Score = 121 bits (304), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 50/77 (64%), Positives = 68/77 (88%), Gaps = 0/77 (0%)
Query 53 YLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLDT 112
YLPLAL+DKC+GS++W+IMK DKE+ GTLRGFDDY+NMVL+DV +Y+F+ +G+K +L+
Sbjct 10 YLPLALIDKCLGSKIWIIMKNDKEITGTLRGFDDYMNMVLEDVVDYSFSPDGVKTTELND 69
Query 113 ILLNGNNVSILVPGGNP 129
L+NGNNV++LVPGG P
Sbjct 70 ALVNGNNVAMLVPGGKP 86
> cel:F28F8.3 lsm-5; LSM Sm-like protein family member (lsm-5);
K12624 U6 snRNA-associated Sm-like protein LSm5
Length=91
Score = 121 bits (303), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 55/80 (68%), Positives = 68/80 (85%), Gaps = 1/80 (1%)
Query 52 SYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLD 111
+ LPL L+DKC+GS++WVIMK DKE+VGTL GFDDYVNMVL+DV EY TA+G + KLD
Sbjct 11 TLLPLELIDKCIGSKIWVIMKNDKEIVGTLTGFDDYVNMVLEDVVEYENTADGKRMTKLD 70
Query 112 TILLNGNNVSILVPGGN-PE 130
TILLNGN++++LVPGG PE
Sbjct 71 TILLNGNHITMLVPGGEGPE 90
> mmu:66373 Lsm5, 2010208O10Rik, 2310034K10Rik; LSM5 homolog,
U6 small nuclear RNA associated (S. cerevisiae); K12624 U6 snRNA-associated
Sm-like protein LSm5
Length=91
Score = 117 bits (294), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 53/80 (66%), Positives = 65/80 (81%), Gaps = 1/80 (1%)
Query 52 SYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLD 111
LPL LVDKC+GSR+ ++MK DKE+VGTL GFDD+VNMVL+DVTE+ T EG + KLD
Sbjct 11 QLLPLELVDKCIGSRIHIVMKSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKLD 70
Query 112 TILLNGNNVSILVPGGN-PE 130
ILLNGNN+++LVPGG PE
Sbjct 71 QILLNGNNITMLVPGGEGPE 90
> mmu:100044729 u6 snRNA-associated Sm-like protein LSm5-like;
K12624 U6 snRNA-associated Sm-like protein LSm5
Length=91
Score = 117 bits (294), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 53/80 (66%), Positives = 65/80 (81%), Gaps = 1/80 (1%)
Query 52 SYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLD 111
LPL LVDKC+GSR+ ++MK DKE+VGTL GFDD+VNMVL+DVTE+ T EG + KLD
Sbjct 11 QLLPLELVDKCIGSRIHIVMKSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKLD 70
Query 112 TILLNGNNVSILVPGGN-PE 130
ILLNGNN+++LVPGG PE
Sbjct 71 QILLNGNNITMLVPGGEGPE 90
> xla:447076 lsm5; LSM5 homolog, U6 small nuclear RNA associated;
K12624 U6 snRNA-associated Sm-like protein LSm5
Length=91
Score = 117 bits (294), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 53/80 (66%), Positives = 65/80 (81%), Gaps = 1/80 (1%)
Query 52 SYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLD 111
LPL LVDKC+GSR+ ++MK DKE+VGTL GFDD+VNMVL+DVTE+ T EG + KLD
Sbjct 11 QLLPLELVDKCIGSRIHIVMKSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKLD 70
Query 112 TILLNGNNVSILVPGGN-PE 130
ILLNGNN+++LVPGG PE
Sbjct 71 QILLNGNNITMLVPGGEGPE 90
> cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM domain
; K12624 U6 snRNA-associated Sm-like protein LSm5
Length=121
Score = 117 bits (293), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 57/95 (60%), Positives = 72/95 (75%), Gaps = 12/95 (12%)
Query 48 SGGPSYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAE---- 103
GG LPLAL+DKC+G+R++V+MKGDKE G LRGFD+YVNMVLDDV EY F A+
Sbjct 22 KGGNIILPLALIDKCIGNRIYVVMKGDKEFSGVLRGFDEYVNMVLDDVQEYGFKADEEDI 81
Query 104 --GIKK------NKLDTILLNGNNVSILVPGGNPE 130
G KK N+L+TILL+GNNV++LVPGG+P+
Sbjct 82 SGGNKKLKRVMVNRLETILLSGNNVAMLVPGGDPD 116
> dre:563948 zgc:171959; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=91
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 55/89 (61%), Positives = 70/89 (78%), Gaps = 2/89 (2%)
Query 44 AGVLSGGPS-YLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTA 102
A + + PS LPL LVDKC+GSR+ ++MK DKE+VGTL GFDD+VNMVL+DVTE+ T
Sbjct 2 AAITATNPSQLLPLELVDKCIGSRIHIVMKNDKEIVGTLLGFDDFVNMVLEDVTEFEITP 61
Query 103 EGIKKNKLDTILLNGNNVSILVPGGN-PE 130
EG + KLD ILLNGNN+++L+PGG PE
Sbjct 62 EGRRITKLDQILLNGNNITMLIPGGEGPE 90
> bbo:BBOV_IV009630 23.m05873; snRNP Sm-like protein; K12624 U6
snRNA-associated Sm-like protein LSm5
Length=87
Score = 115 bits (288), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 47/80 (58%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 47 LSGGPSYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIK 106
+ G YLPLALVDKC+G+++W+IMKG+KE+ G LRGFDDY+N+VLDDVTEYTF G++
Sbjct 4 IESGACYLPLALVDKCLGTKVWIIMKGEKEITGVLRGFDDYMNVVLDDVTEYTFKPTGVE 63
Query 107 KNKLDTILLNGNNVSILVPG 126
+L L+NG N++++VPG
Sbjct 64 TTELKDALVNGTNIAMIVPG 83
> pfa:PF14_0411 small nuclear ribonucleoprotein, putative; K12624
U6 snRNA-associated Sm-like protein LSm5
Length=101
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 51/83 (61%), Positives = 69/83 (83%), Gaps = 0/83 (0%)
Query 47 LSGGPSYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIK 106
+SG ++LPLAL+DKC+GS++W++MKGDKE+VG L GFD+YVNMVL+DVTEYT+ K
Sbjct 4 ISGSETFLPLALMDKCIGSKIWIMMKGDKEIVGKLVGFDEYVNMVLEDVTEYTYANNVKK 63
Query 107 KNKLDTILLNGNNVSILVPGGNP 129
NK+ +LLNG N++I+VPGG P
Sbjct 64 VNKIKKLLLNGLNITIMVPGGTP 86
> hsa:23658 LSM5, FLJ12710, YER146W; LSM5 homolog, U6 small nuclear
RNA associated (S. cerevisiae); K12624 U6 snRNA-associated
Sm-like protein LSm5
Length=62
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 42/61 (68%), Positives = 50/61 (81%), Gaps = 1/61 (1%)
Query 71 MKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLDTILLNGNNVSILVPGGN-P 129
MK DKE+VGTL GFDD+VNMVL+DVTE+ T EG + KLD ILLNGNN+++LVPGG P
Sbjct 1 MKSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKLDQILLNGNNITMLVPGGEGP 60
Query 130 E 130
E
Sbjct 61 E 61
> sce:YER146W LSM5; Lsm (Like Sm) protein; part of heteroheptameric
complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic
Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex
part of U6 snRNP and possibly involved in processing tRNA,
snoRNA, and rRNA; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=93
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 54/83 (65%), Gaps = 5/83 (6%)
Query 51 PSYLPLALVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNK- 109
P LPL ++DK + ++ ++++ ++E GTL GFDD+VN++L+D E+ E +N+
Sbjct 4 PEILPLEVIDKTINQKVLIVLQSNREFEGTLVGFDDFVNVILEDAVEWLIDPEDESRNEK 63
Query 110 ----LDTILLNGNNVSILVPGGN 128
+LL+GNN++ILVPGG
Sbjct 64 VMQHHGRMLLSGNNIAILVPGGK 86
> cel:F40F8.9 lsm-1; LSM Sm-like protein family member (lsm-1);
K12620 U6 snRNA-associated Sm-like protein LSm1
Length=125
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 42/71 (59%), Gaps = 1/71 (1%)
Query 53 YLPLAL-VDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLD 111
YLP A+ + + + +L V+++ ++L+G LR D + N++L+DV E TF + +
Sbjct 7 YLPGAISLFEQLDKKLLVVLRDGRKLIGFLRSIDQFANLILEDVVERTFVEKYFCETGQG 66
Query 112 TILLNGNNVSI 122
+L+ G NV +
Sbjct 67 FMLIRGENVEL 77
> sce:YJL124C LSM1, SPB8; Lsm (Like Sm) protein; forms heteroheptameric
complex (with Lsm2p, Lsm3p, Lsm4p, Lsm5p, Lsm6p, and
Lsm7p) involved in degradation of cytoplasmic mRNAs; K12620
U6 snRNA-associated Sm-like protein LSm1
Length=172
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 50/113 (44%), Gaps = 5/113 (4%)
Query 13 SNSSSINSSNSSSNNSNSNSSSSSSSSSNTAAGVLSGGPSYLPLALVDKCVGSRLWVIMK 72
+NS N SN + N S A + ++ A + V +++V+++
Sbjct 3 ANSKDRNQSNQDAKRQQQNFPKKISEGE---ADLYLDQYNFTTTAAIVSSVDRKIFVLLR 59
Query 73 GDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLDT--ILLNGNNVSIL 123
+ L G LR FD Y N++L D E + +E K + D ++ G NV +L
Sbjct 60 DGRMLFGVLRTFDQYANLILQDCVERIYFSEENKYAEEDRGIFMIRGENVVML 112
> ath:AT4G30330 small nuclear ribonucleoprotein E, putative /
snRNP-E, putative / Sm protein E, putative; K11097 small nuclear
ribonucleoprotein E
Length=88
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 45/78 (57%), Gaps = 6/78 (7%)
Query 55 PLALVDKCVGSR----LWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKL 110
P+ L+ + + S+ +W+ + D + G + GFD+Y+N+VLD+ E + + + L
Sbjct 13 PINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEEVSIKKKT--RKPL 70
Query 111 DTILLNGNNVSILVPGGN 128
ILL G+N+++++ G
Sbjct 71 GRILLKGDNITLMMNAGK 88
> pfa:MAL13P1.253 small nuclear ribonucleoprotein, putative; K11097
small nuclear ribonucleoprotein E
Length=93
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 40/68 (58%), Gaps = 8/68 (11%)
Query 66 RLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKN---KLDTILLNGNNVSI 122
++W+ K D + G + GFD+Y+NMVLD E + +KKN +L ILL G+ +++
Sbjct 30 QIWLYDKPDMRIEGIILGFDEYMNMVLDQTKEIS-----VKKNTKKELGKILLKGDTITL 84
Query 123 LVPGGNPE 130
++ N E
Sbjct 85 IMEVKNEE 92
> tpv:TP03_0386 small nuclear ribonucleoprotein; K11097 small
nuclear ribonucleoprotein E
Length=91
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 41/58 (70%), Gaps = 2/58 (3%)
Query 66 RLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTFTAEGIKKNKLDTILLNGNNVSIL 123
++W+ + + ++ GT+RGFD+Y+NMVLD+ TE + +KK+ + ILL G+ ++++
Sbjct 30 QIWLFDQPNTKIEGTIRGFDEYMNMVLDNATE-VHVKKNVKKD-VGRILLKGDCMTLI 85
> ath:AT1G65700 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12627 U6 snRNA-associated
Sm-like protein LSm8
Length=98
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 40/67 (59%), Gaps = 5/67 (7%)
Query 58 LVDKCVGSRLWVIMKGDKELVGTLRGFDDYVNMVLDDVTEYTF-TAEGIKKNKLDTILLN 116
LVD+ + VI + +VG L+GFD N++LD+ E F T EG++++ L ++
Sbjct 10 LVDQIIS----VITNDGRNIVGVLKGFDQATNIILDESHERVFSTKEGVQQHVLGLYIIR 65
Query 117 GNNVSIL 123
G+N+ ++
Sbjct 66 GDNIGVI 72
> cpv:cgd7_420 protein with DEXDc plus ring plus HELICc; possible
SNF2 domain
Length=2042
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 38/47 (80%), Gaps = 1/47 (2%)
Query 1 STSANNNTS-PNNSNSSSINSSNSSSNNSNSNSSSSSSSSSNTAAGV 46
S +AN+NT+ N SNS++ NS+ S+SN +NSN+++S++S+SNTA +
Sbjct 309 SNTANSNTANSNTSNSNTANSNTSNSNTANSNTANSNTSNSNTANSI 355
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 37/44 (84%), Gaps = 1/44 (2%)
Query 1 STSANNNTS-PNNSNSSSINSSNSSSNNSNSNSSSSSSSSSNTA 43
S +AN+NT+ N +NS++ NS+ ++SN SNSN+++S++S+SNTA
Sbjct 294 SNTANSNTANFNTANSNTANSNTANSNTSNSNTANSNTSNSNTA 337
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 36/44 (81%), Gaps = 1/44 (2%)
Query 1 STSANNNTS-PNNSNSSSINSSNSSSNNSNSNSSSSSSSSSNTA 43
S +AN NT+ N +NS++ NS+ S+SN +NSN+S+S++++SNTA
Sbjct 299 SNTANFNTANSNTANSNTANSNTSNSNTANSNTSNSNTANSNTA 342
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/43 (48%), Positives = 36/43 (83%), Gaps = 4/43 (9%)
Query 1 STSANNNTSPNNSNSSSINSSNSSSNNSNSNSSSSSSSSSNTA 43
S +AN+NT+ NSN+++ N++NS N +NSN+++S++S+SNTA
Sbjct 289 SNTANSNTA--NSNTANFNTANS--NTANSNTANSNTSNSNTA 327
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 37/46 (80%), Gaps = 5/46 (10%)
Query 1 STSANNNTSPNNSNSSSINSSNSSS---NNSNSNSSSSSSSSSNTA 43
S +AN NT+ NSN+++ N++NS++ N +NSN+++S++++SNTA
Sbjct 259 SNTANFNTA--NSNTANFNTANSNTANFNTTNSNTANSNTANSNTA 302
Score = 28.1 bits (61), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 36/42 (85%), Gaps = 1/42 (2%)
Query 3 SANNNTS-PNNSNSSSINSSNSSSNNSNSNSSSSSSSSSNTA 43
+AN+NT+ N +NS++ NS+ ++SN SNSN+++S++++SNT+
Sbjct 306 TANSNTANSNTANSNTSNSNTANSNTSNSNTANSNTANSNTS 347
Lambda K H
0.301 0.121 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40