bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1663_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
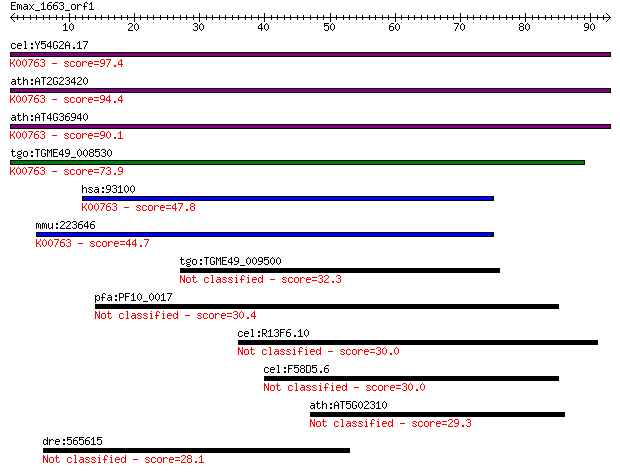
cel:Y54G2A.17 hypothetical protein; K00763 nicotinate phosphor... 97.4 9e-21
ath:AT2G23420 NAPRT2; NAPRT2 (NICOTINATE PHOSPHORIBOSYLTRANSFE... 94.4 8e-20
ath:AT4G36940 NAPRT1; NAPRT1 (NICOTINATE PHOSPHORIBOSYLTRANSFE... 90.1 2e-18
tgo:TGME49_008530 nicotinate phosphoribosyltransferase, putati... 73.9 1e-13
hsa:93100 NAPRT1, PP3856; nicotinate phosphoribosyltransferase... 47.8 9e-06
mmu:223646 Naprt1, 9130210N20Rik; nicotinate phosphoribosyltra... 44.7 7e-05
tgo:TGME49_009500 hypothetical protein 32.3 0.37
pfa:PF10_0017 Plasmodium exported protein (PHISTa) 30.4 1.3
cel:R13F6.10 cra-1; Central Region Assembly in meiosis abnorma... 30.0 2.1
cel:F58D5.6 hypothetical protein 30.0 2.1
ath:AT5G02310 PRT6; PRT6 (PROTEOLYSIS 6); ubiquitin-protein li... 29.3 3.5
dre:565615 Ras and Rab interactor 2-like 28.1 6.7
> cel:Y54G2A.17 hypothetical protein; K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=563
Score = 97.4 bits (241), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 49/94 (52%), Positives = 65/94 (69%), Gaps = 2/94 (2%)
Query 1 LCRHPFQ-ESKRCFVKPKKVEKLLNVWWEKGTITKDLPSLAEIRAHVQTSL-NKVRFDML 58
LCRHPF+ ESKR V K+ KL NV+W+ G + LP+L EI+ HV S+ + +R D
Sbjct 470 LCRHPFEVESKRALVNANKIIKLHNVYWKDGEMITPLPTLNEIKEHVNESIRSTLRQDHR 529
Query 59 RLYNATPYKVSVTDDLYRNTHTLWLENVPIGELQ 92
R N TPYKVSV++ LY+ HTLWL+N PIG+L+
Sbjct 530 RYLNPTPYKVSVSERLYQFLHTLWLQNAPIGQLE 563
> ath:AT2G23420 NAPRT2; NAPRT2 (NICOTINATE PHOSPHORIBOSYLTRANSFERASE
2); nicotinate phosphoribosyltransferase (EC:2.4.2.11);
K00763 nicotinate phosphoribosyltransferase [EC:2.4.2.11]
Length=557
Score = 94.4 bits (233), Expect = 8e-20, Method: Composition-based stats.
Identities = 47/95 (49%), Positives = 59/95 (62%), Gaps = 4/95 (4%)
Query 1 LCRHPFQESKRCFVKPKKVEKLLNVWWEKGTITKD---LPSLAEIRAHVQTSLNKVRFDM 57
LCRHPF ESKR +V P++VE+LL +W +G+ + LP L EIR L +R D
Sbjct 464 LCRHPFNESKRAYVVPQRVEELLKCYW-RGSADEAREVLPPLKEIRDRCIKQLENMRPDH 522
Query 58 LRLYNATPYKVSVTDDLYRNTHTLWLENVPIGELQ 92
+R N TPYKVSV+ LY H LWL P+GELQ
Sbjct 523 MRRLNPTPYKVSVSAKLYDFIHFLWLNEAPVGELQ 557
> ath:AT4G36940 NAPRT1; NAPRT1 (NICOTINATE PHOSPHORIBOSYLTRANSFERASE
1); nicotinate phosphoribosyltransferase (EC:2.4.2.11);
K00763 nicotinate phosphoribosyltransferase [EC:2.4.2.11]
Length=559
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 43/94 (45%), Positives = 55/94 (58%), Gaps = 2/94 (2%)
Query 1 LCRHPFQESKRCFVKPKKVEKLLNVWWEKGT--ITKDLPSLAEIRAHVQTSLNKVRFDML 58
LCRHPF ESKR +V P++VE+LL +W ++L L E+R L +R D +
Sbjct 466 LCRHPFNESKRAYVVPQRVEELLKCYWRGNADEAREELEPLKELRNRCIKQLENMRPDHM 525
Query 59 RLYNATPYKVSVTDDLYRNTHTLWLENVPIGELQ 92
R N TPYKVSV+ LY H LWL P+GEL
Sbjct 526 RRLNPTPYKVSVSAKLYDFIHFLWLNEAPVGELH 559
> tgo:TGME49_008530 nicotinate phosphoribosyltransferase, putative
(EC:2.4.2.11); K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=589
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 32/88 (36%), Positives = 50/88 (56%), Gaps = 0/88 (0%)
Query 1 LCRHPFQESKRCFVKPKKVEKLLNVWWEKGTITKDLPSLAEIRAHVQTSLNKVRFDMLRL 60
CRH + + KRCF+ P KV++LL + G + + SL E R T L +R D+ RL
Sbjct 498 FCRHLYDDRKRCFLVPSKVQRLLQPYISHGKLVVEPLSLEEARMQCITGLRSLRKDLTRL 557
Query 61 YNATPYKVSVTDDLYRNTHTLWLENVPI 88
N TP+KVS +++ + H +W + P+
Sbjct 558 VNPTPFKVSASEEYFTCFHRMWQDTAPV 585
> hsa:93100 NAPRT1, PP3856; nicotinate phosphoribosyltransferase
domain containing 1 (EC:2.4.2.11); K00763 nicotinate phosphoribosyltransferase
[EC:2.4.2.11]
Length=538
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 40/63 (63%), Gaps = 0/63 (0%)
Query 12 CFVKPKKVEKLLNVWWEKGTITKDLPSLAEIRAHVQTSLNKVRFDMLRLYNATPYKVSVT 71
C V+P +VE LL + ++G + + LPSLAE RA Q SL+++ + RL + Y+V ++
Sbjct 463 CTVRPAQVEPLLRLCLQQGQLCEPLPSLAESRALAQLSLSRLSPEHRRLRSPAQYQVVLS 522
Query 72 DDL 74
+ L
Sbjct 523 ERL 525
> mmu:223646 Naprt1, 9130210N20Rik; nicotinate phosphoribosyltransferase
domain containing 1 (EC:2.4.2.11); K00763 nicotinate
phosphoribosyltransferase [EC:2.4.2.11]
Length=538
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 40/70 (57%), Gaps = 0/70 (0%)
Query 5 PFQESKRCFVKPKKVEKLLNVWWEKGTITKDLPSLAEIRAHVQTSLNKVRFDMLRLYNAT 64
P + C VKP +VE LL ++ ++G + + LPSL E R Q SL+ +R +L +
Sbjct 456 PRGTQEPCTVKPAQVEPLLRLYLQQGQLCEPLPSLDESRRFAQQSLSLLRPAHKQLQSPA 515
Query 65 PYKVSVTDDL 74
Y V++++ L
Sbjct 516 VYPVALSEKL 525
> tgo:TGME49_009500 hypothetical protein
Length=2981
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 5/49 (10%)
Query 27 WEKGTITKDLPSLAEIRAHVQTSLNKVRFDMLRLYNATPYKVSVTDDLY 75
W G +TK LP+ EI VQ S + +L+ P K T D+Y
Sbjct 1011 WRDGEVTKVLPASGEIYVRVQAS-----YQDYKLWRLRPPKAHATRDIY 1054
> pfa:PF10_0017 Plasmodium exported protein (PHISTa)
Length=278
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 36/72 (50%), Gaps = 16/72 (22%)
Query 14 VKPKKVEKLLNVWWEKGTITKDLPS-LAEIRAHVQTSLNKVRFDMLRLYNATPYKVSVTD 72
+ PK+V L+N+W++ I KD+ L E++ +V LNK PY D
Sbjct 150 IPPKRV--LINLWYQSLNIAKDMKELLKELKGYVDEYLNK----------NNPYDC---D 194
Query 73 DLYRNTHTLWLE 84
+ + +++W++
Sbjct 195 NFINDNNSIWID 206
> cel:R13F6.10 cra-1; Central Region Assembly in meiosis abnormal
family member (cra-1)
Length=958
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 36 LPSLAEIRAHVQTSLNKVRFDMLRLYNATPYKVSVTDDLYRNTHTLWLENVPIGE 90
+P + H++ S K D++ Y ++ + V DD+ + T TLW + PIGE
Sbjct 580 IPRMTAASKHMKLSAQKTACDVMNRYLSSLF---VLDDVDQITVTLWGDEDPIGE 631
> cel:F58D5.6 hypothetical protein
Length=787
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 28/45 (62%), Gaps = 1/45 (2%)
Query 40 AEIRAHVQTSLNKVRFDMLRLYNATPYKVSVTDDLYRNTHTLWLE 84
A+++AH+ T +N RFD+LR Y+A+ + S T L N+ LE
Sbjct 165 AQLKAHLVTIVNFNRFDLLRNYSASIIQTS-TVALTLNSQKTMLE 208
> ath:AT5G02310 PRT6; PRT6 (PROTEOLYSIS 6); ubiquitin-protein
ligase
Length=2006
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 11/39 (28%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 47 QTSLNKVRFDMLRLYNATPYKVSVTDDLYRNTHTLWLEN 85
Q+ LN V+ ++ +++N P + + D+L R++ +WL++
Sbjct 1803 QSELNHVQ-ELEKMFNIPPIDIILNDELLRSSTQIWLQH 1840
> dre:565615 Ras and Rab interactor 2-like
Length=898
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 11/47 (23%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 6 FQESKRCFVKPKKVEKLLNVWWEKGTITKDLPSLAEIRAHVQTSLNK 52
FQE + + L+ W ++ T+ ++LPS+ + + +++ +L K
Sbjct 763 FQEEQAARILSSATSDTLHQWHQRRTLQRNLPSVDDFQNYMRVALQK 809
Lambda K H
0.321 0.135 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003222032
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40