bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1648_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
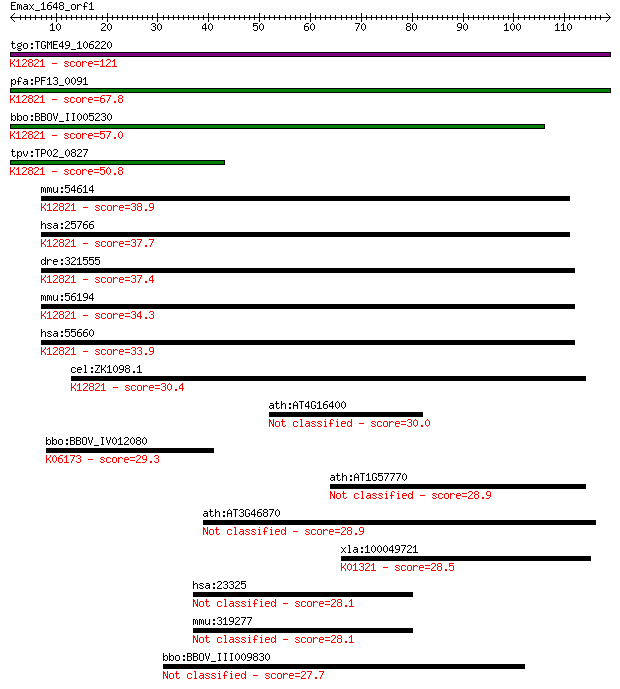
tgo:TGME49_106220 formin binding protein, putative ; K12821 pr... 121 6e-28
pfa:PF13_0091 conserved Plasmodium protein, unknown function; ... 67.8 9e-12
bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12... 57.0 2e-08
tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing... 50.8 1e-06
mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mR... 38.9 0.004
hsa:25766 PRPF40B, HYPC; PRP40 pre-mRNA processing factor 40 h... 37.7 0.008
dre:321555 prpf40a, fb18h07, fe47a12, fnbp3, wu:fb18h07, wu:fe... 37.4 0.012
mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40 ... 34.3 0.11
hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-... 33.9 0.12
cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing ... 30.4 1.3
ath:AT4G16400 hypothetical protein 30.0 2.1
bbo:BBOV_IV012080 23.m06054; tRNA pseudouridine synthase (EC:4... 29.3 3.2
ath:AT1G57770 amine oxidase family 28.9 4.5
ath:AT3G46870 pentatricopeptide (PPR) repeat-containing protein 28.9 4.6
xla:100049721 f9; coagulation factor 9 (EC:3.4.21.22); K01321 ... 28.5 6.0
hsa:23325 KIAA1033 28.1 6.4
mmu:319277 A230046K03Rik, AA589518, Kiaa1033, mKIAA1033; RIKEN... 28.1 6.7
bbo:BBOV_III009830 17.m10622; hypothetical protein 27.7 9.4
> tgo:TGME49_106220 formin binding protein, putative ; K12821
pre-mRNA-processing factor 40
Length=601
Score = 121 bits (303), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 54/118 (45%), Positives = 88/118 (74%), Gaps = 0/118 (0%)
Query 1 LLEWKELSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRRED 60
L W+EL+ TTYI +AD+ H +EWWT++ E ERDDFFQ++M + + ++ K++R++D
Sbjct 262 LQNWEELAPGTTYIAMADKMHEQEWWTFLTEQERDDFFQDYMEEFDKRHRELFKKKRKKD 321
Query 61 VAMLEEILEQNPQEYPFQKKWVDVKDQLLSHPKLQNMMKIDVLQVWEEWVRHGYDQER 118
V +E+IL++ E+ +++KWVDV+D+L + P+L ++++D+LQVWE WV HGY ER
Sbjct 322 VETVEKILDKRSAEFDYRRKWVDVRDELFAIPELSTVLRLDILQVWENWVEHGYADER 379
> pfa:PF13_0091 conserved Plasmodium protein, unknown function;
K12821 pre-mRNA-processing factor 40
Length=906
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 42/122 (34%), Positives = 69/122 (56%), Gaps = 8/122 (6%)
Query 1 LLEWKELSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRRED 60
LL W +L+ TTY++ A +F+ +EWW W+ E ERD+ FQ++M K FK+ +++R++
Sbjct 517 LLNWDKLNECTTYVEFASQFYKQEWWEWITEKERDEVFQDFMDGYKSKFKETRRKKRKQK 576
Query 61 VAMLEEILEQNPQEYPFQK----KWVDVKDQLLSHPKLQNMMKIDVLQVWEEWVRHGYDQ 116
+ EIL+Q QEY KW DV+ ++ KID L WE++ ++
Sbjct 577 M----EILKQKFQEYATDNKNPLKWNDVQKYFRDDEDFHSLHKIDALAAWEDFYEKYHNV 632
Query 117 ER 118
E+
Sbjct 633 EK 634
> bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12821
pre-mRNA-processing factor 40
Length=457
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 57/106 (53%), Gaps = 7/106 (6%)
Query 1 LLEWKELSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRRED 60
L +W+EL+ TTY++ A+R H EWWTW E RD FQE M K++ ++RRR
Sbjct 173 LEKWEELTPYTTYVEFAERCHTREWWTWADEKTRDGIFQETMERMDHELKERQRERRRVS 232
Query 61 VAMLEEILEQ-NPQEYPFQKKWVDVKDQLLSHPKLQNMMKIDVLQV 105
+ LE +E+ + P +W +VK Q + + +D+L+
Sbjct 233 MEKLEAEMEKLVTDDMP---QWPNVKRQFTG---FEGLHLLDILEC 272
> tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=390
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 1 LLEWKELSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWM 42
LL W+ELS+ T Y D + +FH EWW W E RD FQE+M
Sbjct 171 LLNWEELSYATVYADFSKQFHTAEWWDWGDEVTRDAIFQEFM 212
> mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mRNA
processing factor 40 homolog B (yeast); K12821 pre-mRNA-processing
factor 40
Length=873
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 51/110 (46%), Gaps = 7/110 (6%)
Query 7 LSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRREDVAMLEE 66
++ T Y F E W + E ER + + + +F + K++ KQ RR ++ L+
Sbjct 360 MTSTTRYRRAEQTFGDLEVWAVVPERERKEVYDDVLFFLAKKEKEQAKQLRRRNIQALKS 419
Query 67 ILEQNPQEYPFQKKWVDVKDQLLSHP------KLQNMMKIDVLQVWEEWV 110
IL+ FQ W + L+ +P +LQNM K D L +EE +
Sbjct 420 ILD-GMSSVNFQTTWSQAQQYLMDNPSFAQDQQLQNMDKEDALICFEEHI 468
> hsa:25766 PRPF40B, HYPC; PRP40 pre-mRNA processing factor 40
homolog B (S. cerevisiae); K12821 pre-mRNA-processing factor
40
Length=871
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 51/110 (46%), Gaps = 7/110 (6%)
Query 7 LSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRREDVAMLEE 66
++ T Y F E W + E +R + + + +F + K++ KQ RR ++ L+
Sbjct 360 MTSTTRYRRAEQTFGELEVWAVVPERDRKEVYDDVLFFLAKKEKEQAKQLRRRNIQALKS 419
Query 67 ILEQNPQEYPFQKKWVDVKDQLLSHP------KLQNMMKIDVLQVWEEWV 110
IL+ FQ W + L+ +P +LQNM K D L +EE +
Sbjct 420 ILD-GMSSVNFQTTWSQAQQYLMDNPSFAQDHQLQNMDKEDALICFEEHI 468
> dre:321555 prpf40a, fb18h07, fe47a12, fnbp3, wu:fb18h07, wu:fe47a12;
PRP40 pre-mRNA processing factor 40 homolog A (yeast);
K12821 pre-mRNA-processing factor 40
Length=851
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 56/111 (50%), Gaps = 7/111 (6%)
Query 7 LSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRREDVAMLEE 66
++ T Y F +E W+ + E +R + +++ +F + K++ KQ R+ + L+
Sbjct 371 MTSTTRYKKAEQMFGDQEVWSCVPERDRLEIYEDVLFYLAKKEKEQAKQLRKRNWEALKN 430
Query 67 ILEQNPQEYPFQKKWVDVKDQLLSHP------KLQNMMKIDVLQVWEEWVR 111
IL+ N ++ W + + LL +P +LQNM K D L +EE +R
Sbjct 431 ILD-NMANVTYRTTWSEAQQYLLDNPTFAEDEELQNMDKEDALICFEEHIR 480
> mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40
pre-mRNA processing factor 40 homolog A (yeast); K12821 pre-mRNA-processing
factor 40
Length=953
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 53/111 (47%), Gaps = 7/111 (6%)
Query 7 LSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRREDVAMLEE 66
++ T Y F E W + E +R + +++ +F + K++ KQ R+ + L+
Sbjct 473 MTSTTRYKKAEQMFGEMEVWNAISERDRLEIYEDVLFFLSKKEKEQAKQLRKRNWEALKN 532
Query 67 ILEQNPQEYPFQKKWVDVKDQLLSHP------KLQNMMKIDVLQVWEEWVR 111
IL+ N + W + + L+ +P +LQNM K D L +EE +R
Sbjct 533 ILD-NMANVTYSTTWSEAQQYLMDNPTFAEDEELQNMDKEDALICFEEHIR 582
> hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-10,
HIP10, HYPA, NY-REN-6, Prp40; PRP40 pre-mRNA processing
factor 40 homolog A (S. cerevisiae); K12821 pre-mRNA-processing
factor 40
Length=930
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 53/111 (47%), Gaps = 7/111 (6%)
Query 7 LSFKTTYIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRREDVAMLEE 66
++ T Y F E W + E +R + +++ +F + K++ KQ R+ + L+
Sbjct 450 MTSTTRYKKAEQMFGEMEVWNAISERDRLEIYEDVLFFLSKKEKEQAKQLRKRNWEALKN 509
Query 67 ILEQNPQEYPFQKKWVDVKDQLLSHP------KLQNMMKIDVLQVWEEWVR 111
IL+ N + W + + L+ +P +LQNM K D L +EE +R
Sbjct 510 ILD-NMANVTYSTTWSEAQQYLMDNPTFAEDEELQNMDKEDALICFEEHIR 559
> cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=724
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/107 (22%), Positives = 51/107 (47%), Gaps = 7/107 (6%)
Query 13 YIDVADRFHLEEWWTWMQESERDDFFQEWMFDNKQMFKDKIKQRRREDVAMLEEILEQNP 72
Y +D F E W + + +R + F++ + + K+K ++ R+ D+A +L Q+
Sbjct 318 YQKASDIFSKEPLWIAVNDEDRKEIFRDCIDFVARRDKEKKEEDRKRDIAAFSHVL-QSM 376
Query 73 QEYPFQKKWVDVKDQLLSHPK------LQNMMKIDVLQVWEEWVRHG 113
++ ++ W + L +P+ L M K D L V+E+ ++
Sbjct 377 EQITYKTTWAQAQRILYENPQFAERKDLHFMDKEDALTVFEDHIKQA 423
> ath:AT4G16400 hypothetical protein
Length=218
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 20/30 (66%), Gaps = 4/30 (13%)
Query 52 KIKQRRREDVAMLEEILEQNPQEYPFQKKW 81
KI R++ LE++L+QNPQ FQ+KW
Sbjct 50 KITLTRKQ----LEQLLQQNPQGVSFQRKW 75
> bbo:BBOV_IV012080 23.m06054; tRNA pseudouridine synthase (EC:4.2.1.70);
K06173 tRNA pseudouridine synthase A [EC:5.4.99.12]
Length=414
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 8 SFKTT--YIDVADRFHLEEWWTWMQESERDDFFQE 40
+FK T Y ++A F + W TWM++ R+ F+ E
Sbjct 358 NFKRTRIYPEIASTFETDVWKTWMEKLLRNPFYLE 392
> ath:AT1G57770 amine oxidase family
Length=574
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 12/50 (24%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 64 LEEILEQNPQEYPFQKKWVDVKDQLLSHPKLQNMMKIDVLQVWEEWVRHG 113
EI++ + PF + W+D+ LL+ K ++ +++ ++ EW + G
Sbjct 236 FSEIVDSLELKDPFIRNWIDLLAFLLAGVKSDGILSAEMIYMFAEWYKPG 285
> ath:AT3G46870 pentatricopeptide (PPR) repeat-containing protein
Length=257
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 45/84 (53%), Gaps = 13/84 (15%)
Query 39 QEWMFDNKQMFKDKIKQRRREDVA-----MLEEILEQN--PQEYPFQKKWVDVKDQLLSH 91
+E +F + Q + + I+ R+ + E++L+ P+E PF+ V +K LL H
Sbjct 171 KENLFPDSQTYTEVIRGFLRDGCPADAMNVYEDMLKSPDPPEELPFR---VLLKG-LLPH 226
Query 92 PKLQNMMKIDVLQVWEEWVRHGYD 115
P L+N +K D +++ E +H YD
Sbjct 227 PLLRNKVKKDFEELFPE--KHAYD 248
> xla:100049721 f9; coagulation factor 9 (EC:3.4.21.22); K01321
coagulation factor IX (Christmas factor) [EC:3.4.21.22]
Length=461
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 66 EILEQNPQEYPFQKKWVDVKDQLLSHPKLQNMMKIDVLQVWEEWVRHGY 114
E+ E + + F K+++D DQ LS+P L D + + W + GY
Sbjct 69 EVFENDDKTKEFWKQYID-GDQCLSNPCLNGGACKDDVSAYVCWCQQGY 116
> hsa:23325 KIAA1033
Length=1173
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query 37 FFQEWMFDNKQMFKDKIKQRRREDVAMLEEILEQNPQEYPFQK 79
F ++M+D + IK R +D+ EI +QN +YPF +
Sbjct 852 IFSQFMYD------EHIKSRLIKDIRFFREIKDQNDHKYPFDR 888
> mmu:319277 A230046K03Rik, AA589518, Kiaa1033, mKIAA1033; RIKEN
cDNA A230046K03 gene
Length=1173
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query 37 FFQEWMFDNKQMFKDKIKQRRREDVAMLEEILEQNPQEYPFQK 79
F ++M+D + IK R +D+ EI +QN +YPF +
Sbjct 852 IFSQFMYD------EHIKSRLIKDIRFFREIKDQNDHKYPFDR 888
> bbo:BBOV_III009830 17.m10622; hypothetical protein
Length=1593
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 38/78 (48%), Gaps = 12/78 (15%)
Query 31 ESERDDF-------FQEWMFDNKQMFKDKIKQRRREDVAMLEEILEQNPQEYPFQKKWVD 83
+SE +D+ F+ + NK+ +K++QR V L ++ +P KK++
Sbjct 291 QSELEDYRTHSTLVFKRYRLSNKECSLEKVEQRINNAVITLRTLVT-----FPLTKKYIP 345
Query 84 VKDQLLSHPKLQNMMKID 101
Q+L + +++M +D
Sbjct 346 GLKQMLESNEYEDLMDMD 363
Lambda K H
0.321 0.135 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40