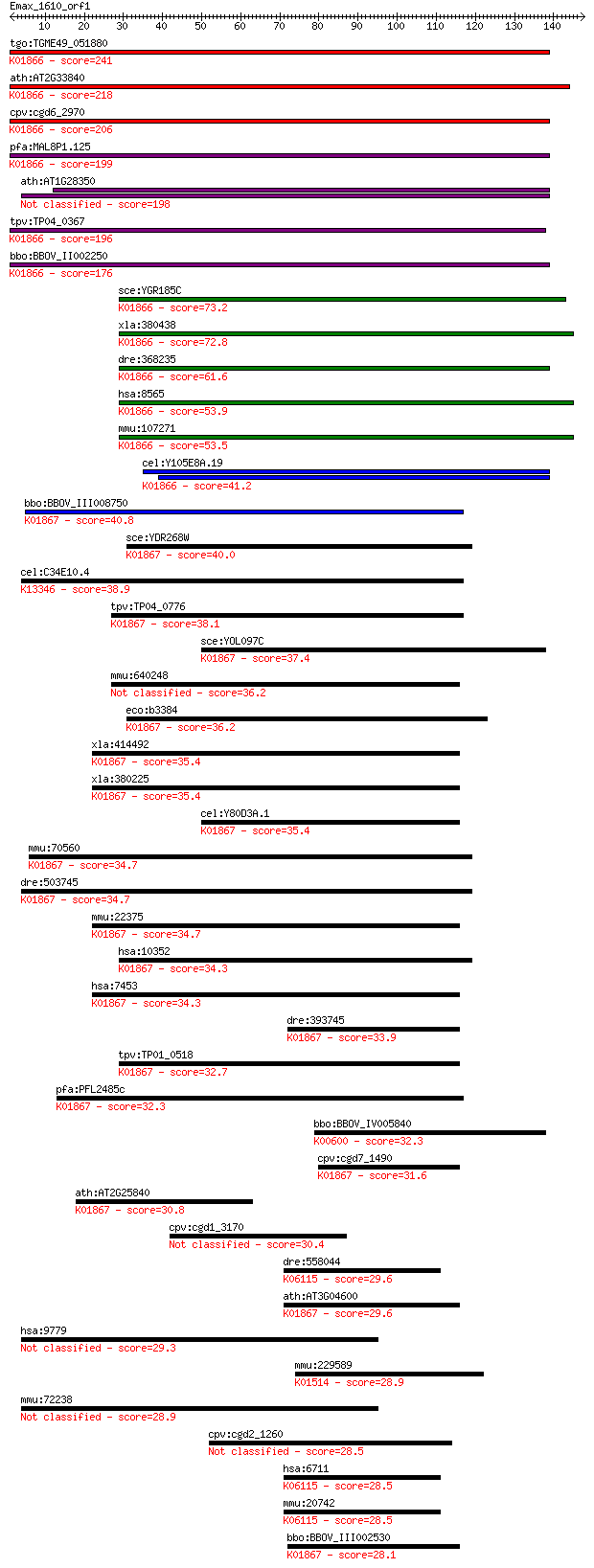
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1610_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_051880 tyrosyl-tRNA synthetase, putative (EC:6.1.1.... 241 4e-64
ath:AT2G33840 tRNA synthetase class I (W and Y) family protein... 218 7e-57
cpv:cgd6_2970 tyrosyl-tRNA synthetase (tyrosyl-tRNA ligase; Ty... 206 3e-53
pfa:MAL8P1.125 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);... 199 3e-51
ath:AT1G28350 ATP binding / aminoacyl-tRNA ligase/ nucleotide ... 198 4e-51
tpv:TP04_0367 tyrosyl-tRNA synthetase (EC:6.1.1.1); K01866 tyr... 196 2e-50
bbo:BBOV_II002250 18.m06182; tyrosyl-tRNA synthetase; K01866 t... 176 2e-44
sce:YGR185C TYS1, TTS1; Cytoplasmic tyrosyl-tRNA synthetase, r... 73.2 3e-13
xla:380438 yars, MGC53284; tyrosyl-tRNA synthetase (EC:6.1.1.1... 72.8 4e-13
dre:368235 yars, cb204, wu:fk52b08; tyrosyl-tRNA synthetase (E... 61.6 9e-10
hsa:8565 YARS, CMTDIC, TYRRS, YRS, YTS; tyrosyl-tRNA synthetas... 53.9 2e-07
mmu:107271 Yars, AL024047, AU018965; tyrosyl-tRNA synthetase (... 53.5 2e-07
cel:Y105E8A.19 hypothetical protein; K01866 tyrosyl-tRNA synth... 41.2 0.001
bbo:BBOV_III008750 17.m07764; tRNA synthetases class I (W and ... 40.8 0.002
sce:YDR268W MSW1; Msw1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA... 40.0 0.003
cel:C34E10.4 wrs-2; tryptophanyl (W) tRNA Synthetase family me... 38.9 0.006
tpv:TP04_0776 tryptophanyl-tRNA synthetase (EC:6.1.1.2); K0186... 38.1 0.009
sce:YOL097C WRS1, HRE342; Wrs1p (EC:6.1.1.2); K01867 tryptopha... 37.4 0.019
mmu:640248 tryptophanyl-tRNA synthetase, cytoplasmic-like 36.2 0.037
eco:b3384 trpS, ECK3371, JW3347; tryptophanyl-tRNA synthetase ... 36.2 0.041
xla:414492 wars, MGC81110, gamma-2, ifi53, ifp53; tryptophanyl... 35.4 0.058
xla:380225 wars, MGC53671; tryptophanyl-tRNA synthetase (EC:6.... 35.4 0.064
cel:Y80D3A.1 wrs-1; tryptophanyl (W) tRNA Synthetase family me... 35.4 0.069
mmu:70560 Wars2, 5730427B17Rik, 9430020O07Rik, AI413375, TrpRS... 34.7 0.11
dre:503745 wars2, zgc:110828; tryptophanyl tRNA synthetase 2, ... 34.7 0.11
mmu:22375 Wars, TrpRS, WRS; tryptophanyl-tRNA synthetase (EC:6... 34.7 0.12
hsa:10352 WARS2, TrpRS; tryptophanyl tRNA synthetase 2, mitoch... 34.3 0.14
hsa:7453 WARS, GAMMA-2, IFI53, IFP53; tryptophanyl-tRNA synthe... 34.3 0.15
dre:393745 wars, MGC73274, zgc:73274; tryptophanyl-tRNA synthe... 33.9 0.17
tpv:TP01_0518 tryptophanyl-tRNA synthetase; K01867 tryptophany... 32.7 0.39
pfa:PFL2485c tryptophanyl-tRNA synthetase, putative (EC:6.1.1.... 32.3 0.54
bbo:BBOV_IV005840 23.m06426; serine hydroxymethyltransferase (... 32.3 0.57
cpv:cgd7_1490 tryptophanyl-tRNA synthetase ; K01867 tryptophan... 31.6 0.96
ath:AT2G25840 OVA4; OVA4 (ovule abortion 4); ATP binding / ami... 30.8 1.7
cpv:cgd1_3170 hypothetical protein 30.4 1.8
dre:558044 spectrin, beta, non-erythrocytic 1; K06115 spectrin... 29.6 3.2
ath:AT3G04600 tRNA synthetase class I (W and Y) family protein... 29.6 3.6
hsa:9779 TBC1D5, KIAA0210; TBC1 domain family, member 5 29.3
mmu:229589 Prune, 9230112O05Rik, C130058A12, DRES-17, HTCD37, ... 28.9 6.4
mmu:72238 Tbc1d5, 1600014N05Rik, KIAA0210, mKIAA0210; TBC1 dom... 28.9 6.6
cpv:cgd2_1260 hypothetical protein 28.5 6.9
hsa:6711 SPTBN1, ELF, SPTB2, betaSpII; spectrin, beta, non-ery... 28.5 8.6
mmu:20742 Spnb2, 9930031C03Rik, AL033301, KIAA4049, SPTB2, SPT... 28.5 8.7
bbo:BBOV_III002530 17.m07244; tryptophanyl-tRNA synthetase fam... 28.1 9.5
> tgo:TGME49_051880 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=427
Score = 241 bits (616), Expect = 4e-64, Method: Compositional matrix adjust.
Identities = 110/138 (79%), Positives = 125/138 (90%), Gaps = 0/138 (0%)
Query 1 AKANSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLA 60
A++ +I+RVKRCSQIMGR EGD+QP+AQIMYPCMQCADIFYL ADICQLGMDQRKVN+LA
Sbjct 212 ARSFTITRVKRCSQIMGRQEGDEQPAAQIMYPCMQCADIFYLGADICQLGMDQRKVNMLA 271
Query 61 REYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPP 120
REYCD IK+K K VILSH MLPGLLEGQ+KMSKSN DSAIFMED+E EVNRKIKKAFCP
Sbjct 272 REYCDEIKRKLKPVILSHQMLPGLLEGQEKMSKSNPDSAIFMEDTETEVNRKIKKAFCPA 331
Query 121 CIIEGNPCISYAEYLVYP 138
+I+GNPC++Y E LV+P
Sbjct 332 GVIDGNPCVTYVEQLVFP 349
> ath:AT2G33840 tRNA synthetase class I (W and Y) family protein;
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=385
Score = 218 bits (554), Expect = 7e-57, Method: Compositional matrix adjust.
Identities = 100/145 (68%), Positives = 122/145 (84%), Gaps = 2/145 (1%)
Query 1 AKANSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLA 60
A+ N + R+ RC QIMGR+E D+ +AQI+YPCMQCADIF+L ADICQLGMDQRKVNVLA
Sbjct 170 ARKNKLPRILRCVQIMGRSETDELSAAQILYPCMQCADIFFLEADICQLGMDQRKVNVLA 229
Query 61 REYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPP 120
REYCD IK+KNK +ILSHHMLPGL +GQ+KMSKS+ SAIFMED EAEVN KIKKA+CPP
Sbjct 230 REYCDDIKRKNKPIILSHHMLPGLQQGQEKMSKSDPLSAIFMEDEEAEVNVKIKKAYCPP 289
Query 121 CIIEGNPCISYAEYLVYP--DEYLV 143
+++GNPC+ Y +Y++ P DE+ V
Sbjct 290 KVVKGNPCLEYIKYIILPWFDEFTV 314
> cpv:cgd6_2970 tyrosyl-tRNA synthetase (tyrosyl-tRNA ligase;
TyrRS). class-I aaRS ; K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=376
Score = 206 bits (523), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 97/138 (70%), Positives = 112/138 (81%), Gaps = 0/138 (0%)
Query 1 AKANSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLA 60
++ +I+R+KRC QIMGR E D+QP A + YPCMQCADIF L ADICQLGMDQRKVN+LA
Sbjct 161 SRKFNITRIKRCCQIMGRQENDEQPCASVFYPCMQCADIFQLKADICQLGMDQRKVNMLA 220
Query 61 REYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPP 120
REYCD K+K VILSH MLPGLLEGQ+KMSKS+T SAIF+ED+ V +KIKKAFCPP
Sbjct 221 REYCDAAGIKHKPVILSHKMLPGLLEGQEKMSKSDTSSAIFVEDTPEAVVKKIKKAFCPP 280
Query 121 CIIEGNPCISYAEYLVYP 138
IIEGNPCI Y LV+P
Sbjct 281 GIIEGNPCIEYINTLVFP 298
> pfa:MAL8P1.125 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=373
Score = 199 bits (506), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 92/138 (66%), Positives = 120/138 (86%), Gaps = 0/138 (0%)
Query 1 AKANSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLA 60
+++ +I+R+KRC +IMGR+EG++ +QI+YPCMQCADIF+LN DICQLG+DQRKVN+LA
Sbjct 158 SRSFNINRMKRCLKIMGRSEGEENYCSQILYPCMQCADIFFLNVDICQLGIDQRKVNMLA 217
Query 61 REYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPP 120
REYCD K K K VILSH MLPGLLEGQ+KMSKS+ +SAIFM+DSE++VNRKIKKA+CPP
Sbjct 218 REYCDIKKIKKKPVILSHGMLPGLLEGQEKMSKSDENSAIFMDDSESDVNRKIKKAYCPP 277
Query 121 CIIEGNPCISYAEYLVYP 138
+IE NP +YA+ +++P
Sbjct 278 NVIENNPIYAYAKSIIFP 295
> ath:AT1G28350 ATP binding / aminoacyl-tRNA ligase/ nucleotide
binding / tyrosine-tRNA ligase
Length=776
Score = 198 bits (504), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 90/127 (70%), Positives = 106/127 (83%), Gaps = 0/127 (0%)
Query 12 CSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKN 71
C QIMGR+E + +AQI+YPCMQCADIF L ADICQLGMDQRKVN+LAREYC IK+KN
Sbjct 573 CGQIMGRSETEVLSAAQILYPCMQCADIFLLEADICQLGMDQRKVNMLAREYCADIKRKN 632
Query 72 KQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISY 131
K +ILSHHMLPGL +GQ+KMSKS+ SAIFMED EA+VN KI KA+CPP +EGNPC+ Y
Sbjct 633 KPIILSHHMLPGLQQGQEKMSKSDPSSAIFMEDEEADVNEKISKAYCPPKTVEGNPCLEY 692
Query 132 AEYLVYP 138
+Y+V P
Sbjct 693 VKYIVLP 699
Score = 197 bits (500), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 87/135 (64%), Positives = 112/135 (82%), Gaps = 0/135 (0%)
Query 4 NSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREY 63
NS++++KRC IMG +E ++ +A I+Y CMQCAD F+L ADICQLGMDQ+ VN+LAR+Y
Sbjct 187 NSLAQIKRCMPIMGLSENEELSAAHILYVCMQCADTFFLEADICQLGMDQQTVNLLARDY 246
Query 64 CDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCII 123
CD +K++NK VILSHHMLPGL +GQ KMSKS+ SAIFMED EAEVN KIKKA+CPP I+
Sbjct 247 CDVVKRENKPVILSHHMLPGLQQGQKKMSKSDPSSAIFMEDEEAEVNVKIKKAYCPPDIV 306
Query 124 EGNPCISYAEYLVYP 138
EGNPC+ Y ++++ P
Sbjct 307 EGNPCLEYVKHIILP 321
> tpv:TP04_0367 tyrosyl-tRNA synthetase (EC:6.1.1.1); K01866 tyrosyl-tRNA
synthetase [EC:6.1.1.1]
Length=427
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 87/137 (63%), Positives = 115/137 (83%), Gaps = 0/137 (0%)
Query 1 AKANSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLA 60
+++ +I+R+KRCSQ +GR EGDDQPSAQ++YP +QCADIF++ ADICQLGMDQRK+NVLA
Sbjct 210 SRSFNITRMKRCSQALGRTEGDDQPSAQLLYPAIQCADIFFIGADICQLGMDQRKINVLA 269
Query 61 REYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPP 120
REY + K V+LSH MLPGL+EGQ+KMSKSN +SAIFM+DS EV+ KIKKA+CPP
Sbjct 270 REYSELKKIPRSPVVLSHRMLPGLVEGQEKMSKSNPNSAIFMDDSPEEVSSKIKKAYCPP 329
Query 121 CIIEGNPCISYAEYLVY 137
+++GNP I+Y ++V+
Sbjct 330 GVVDGNPVIAYFCFIVF 346
> bbo:BBOV_II002250 18.m06182; tyrosyl-tRNA synthetase; K01866
tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=418
Score = 176 bits (446), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 79/138 (57%), Positives = 108/138 (78%), Gaps = 0/138 (0%)
Query 1 AKANSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLA 60
+++ +I+R+KRCS+ +GRA+GD++P A ++YP MQCADIFY+ ADICQLG+DQRK+N+LA
Sbjct 192 SRSFNITRLKRCSEALGRADGDNRPGASLLYPAMQCADIFYIGADICQLGLDQRKINMLA 251
Query 61 REYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPP 120
REYC+ + +ILSHHM+P L + KMSKS +SAIFMEDS EVN KIK A+CP
Sbjct 252 REYCELKSVGARPIILSHHMMPSLAQQGGKMSKSIPNSAIFMEDSAEEVNAKIKSAWCPE 311
Query 121 CIIEGNPCISYAEYLVYP 138
+ NPCI+Y E++V+P
Sbjct 312 KEVCDNPCIAYFEHIVFP 329
> sce:YGR185C TYS1, TTS1; Cytoplasmic tyrosyl-tRNA synthetase,
required for cytoplasmic protein synthesis; interacts with
positions 34 and 35 of the tRNATyr anticodon; mutations in human
ortholog YARS are associated with Charcot-Marie-Tooth (CMT)
neuropathies (EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase
[EC:6.1.1.1]
Length=394
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/115 (39%), Positives = 69/115 (60%), Gaps = 4/115 (3%)
Query 29 IMYPCMQCADIFYLNADICQLG-MDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEG 87
++YP MQ D +L+ D CQ G +DQRK+ VLA E + K K+ L + M+PGL +G
Sbjct 168 LIYPLMQALDEQFLDVD-CQFGGVDQRKIFVLAEENLPSLGYK-KRAHLMNPMVPGLAQG 225
Query 88 QDKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYPDEYL 142
KMS S+ +S I + + +V +KI AFC P +E N +S+ +Y++ P + L
Sbjct 226 -GKMSASDPNSKIDLLEEPKQVKKKINSAFCSPGNVEENGLLSFVQYVIAPIQEL 279
> xla:380438 yars, MGC53284; tyrosyl-tRNA synthetase (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=528
Score = 72.8 bits (177), Expect = 4e-13, Method: Composition-based stats.
Identities = 43/118 (36%), Positives = 69/118 (58%), Gaps = 5/118 (4%)
Query 29 IMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQ 88
++YP +Q D YL D G+DQRK+ A +Y + K++ L + M+PGL
Sbjct 164 LLYPGLQALDEEYLKVDAQFGGVDQRKIFTFAEKYLPALGYA-KRIHLMNPMVPGLTGA- 221
Query 89 DKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYP--DEYLVI 144
KMS S +S I + DS A+V +K+KKAFC P +E N +S+ ++++P E++V+
Sbjct 222 -KMSSSEEESKIDLLDSPADVKKKLKKAFCEPGNVENNGVLSFVRHVLFPLKSEFVVL 278
> dre:368235 yars, cb204, wu:fk52b08; tyrosyl-tRNA synthetase
(EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=529
Score = 61.6 bits (148), Expect = 9e-10, Method: Composition-based stats.
Identities = 40/110 (36%), Positives = 63/110 (57%), Gaps = 3/110 (2%)
Query 29 IMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQ 88
++YP +Q D YL D G+DQRK+ LA +Y + K+ L + M+PGL
Sbjct 164 LLYPGLQALDEEYLKVDAQFGGVDQRKIFTLAEKYLPSLGY-TKRSHLMNPMVPGLT--G 220
Query 89 DKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYP 138
KMS S +S I + D +V +K+KKAFC P +E N +S+ +++++P
Sbjct 221 SKMSSSEEESKIDLLDKNQDVKKKLKKAFCEPGNVENNGVLSFVKHVLFP 270
> hsa:8565 YARS, CMTDIC, TYRRS, YRS, YTS; tyrosyl-tRNA synthetase
(EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=528
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 41/118 (34%), Positives = 70/118 (59%), Gaps = 5/118 (4%)
Query 29 IMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQ 88
++YP +Q D YL D G+DQRK+ A +Y + +K+V L + M+PGL
Sbjct 164 LLYPGLQALDEEYLKVDAQFGGIDQRKIFTFAEKYLPALGY-SKRVHLMNPMVPGLT--G 220
Query 89 DKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYP--DEYLVI 144
KMS S +S I + D + +V +K+KKAFC P +E N +S+ +++++P E++++
Sbjct 221 SKMSSSEEESKIDLLDRKEDVKKKLKKAFCEPGNVENNGVLSFIKHVLFPLKSEFVIL 278
> mmu:107271 Yars, AL024047, AU018965; tyrosyl-tRNA synthetase
(EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=528
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 41/118 (34%), Positives = 70/118 (59%), Gaps = 5/118 (4%)
Query 29 IMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQ 88
++YP +Q D YL D G+DQRK+ A +Y + +K+V L + M+PGL
Sbjct 164 LLYPGLQALDEEYLKVDAQFGGVDQRKIFTFAEKYLPALGY-SKRVHLMNPMVPGLT--G 220
Query 89 DKMSKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYP--DEYLVI 144
KMS S +S I + D + +V +K+KKAFC P +E N +S+ +++++P E++++
Sbjct 221 SKMSSSEEESKIDLLDRKEDVKKKLKKAFCEPGNVENNGVLSFIKHVLFPLKSEFVIL 278
> cel:Y105E8A.19 hypothetical protein; K01866 tyrosyl-tRNA synthetase
[EC:6.1.1.1]
Length=722
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 36/104 (34%), Positives = 52/104 (50%), Gaps = 4/104 (3%)
Query 35 QCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKS 94
Q D YL D G+DQRK+ +LA E + K K+ L + M+PGL KMS S
Sbjct 181 QALDEQYLKVDGQFGGVDQRKIFILAEEQLPKL-KLGKRWHLMNPMVPGLT--GTKMSSS 237
Query 95 NTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYP 138
DS I + D +V KI A C + N +S+ ++++P
Sbjct 238 EEDSKIDVLDEPEKVRSKIMGAACSRDQPD-NGVLSFYNFVLFP 280
Score = 38.9 bits (89), Expect = 0.005, Method: Composition-based stats.
Identities = 32/107 (29%), Positives = 55/107 (51%), Gaps = 12/107 (11%)
Query 39 IFYLNA-----DICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGL--LEGQDKM 91
I+ LNA DI +G D + V A + C + + ++H ++P + GQ KM
Sbjct 533 IYALNAQMTRPDILIIGADSQ---VFA-DLCSRLLRSFGYPAIAHLLIPTIPGCNGQ-KM 587
Query 92 SKSNTDSAIFMEDSEAEVNRKIKKAFCPPCIIEGNPCISYAEYLVYP 138
S S D + D+ ++ KI ++FC P ++GN + AE++V+P
Sbjct 588 SCSIPDFLLDPLDTPKQLKTKIARSFCEPQNLDGNVAMQLAEHIVFP 634
> bbo:BBOV_III008750 17.m07764; tRNA synthetases class I (W and
Y) family protein; K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=388
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 56/122 (45%), Gaps = 10/122 (8%)
Query 5 SISRVKRCSQIMGRAEGDDQPS-AQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREY 63
SI R+++ + R D S A+ +YP + AD+ +D G DQ + RE
Sbjct 159 SIGRLRKLANFEHRQVAQDGDSVAEFLYPLLMAADVLCSGSDSIIAGEDQLSHVCVIREV 218
Query 64 CDYIKK-KNKQVILSHHMLPGL------LEGQDKMSKSNTD--SAIFMEDSEAEVNRKIK 114
+ + V+ MLP L+G+ KMSKS+ S I + D+E + +KIK
Sbjct 219 AKKLNRLAGSTVVCIPDMLPNTGFRVISLDGRGKMSKSSLQACSRINITDTEETIYQKIK 278
Query 115 KA 116
A
Sbjct 279 VA 280
> sce:YDR268W MSW1; Msw1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=379
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 43/98 (43%), Gaps = 10/98 (10%)
Query 31 YPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKKNKQ--------VILSHHMLP 82
YP +Q ADI + +G DQ + L R + K K+ ++ +
Sbjct 177 YPVLQAADILLYKSTHVPVGDDQSQHLELTRHLAEKFNKMYKKNFFPKPVTMLAQTKKVL 236
Query 83 GLLEGQDKMSKS--NTDSAIFMEDSEAEVNRKIKKAFC 118
L + KMSKS N DS IF+ D + +KI+KA
Sbjct 237 SLSTPEKKMSKSDPNHDSVIFLNDEPKAIQKKIRKALT 274
> cel:C34E10.4 wrs-2; tryptophanyl (W) tRNA Synthetase family
member (wrs-2); K13346 peroxin-10
Length=650
Score = 38.9 bits (89), Expect = 0.006, Method: Composition-based stats.
Identities = 35/124 (28%), Positives = 58/124 (46%), Gaps = 11/124 (8%)
Query 4 NSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRK-VNVLA-- 60
+ ++R+ + + R + D P + YP +Q AD+ A +G DQ + +N+L
Sbjct 424 SKLARLPQYKEKKERFKKGDIPVGLLTYPLLQAADVLTFKATTVPVGEDQSQHLNLLGGL 483
Query 61 -----REYCDYIKKKNKQVIL-SHHMLPGLLEGQDKMSKSN--TDSAIFMEDSEAEVNRK 112
+ Y I KQ+ SH + L E + KMSKS+ S I + DS + + K
Sbjct 484 AYAFNKTYETEIFPIPKQLTRESHARIRSLREPEKKMSKSSGGPRSRIEITDSRSTIIEK 543
Query 113 IKKA 116
+KA
Sbjct 544 CQKA 547
> tpv:TP04_0776 tryptophanyl-tRNA synthetase (EC:6.1.1.2); K01867
tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=306
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 50/104 (48%), Gaps = 18/104 (17%)
Query 27 AQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKK-NKQVILSHHMLPGLL 85
A +YP + +DIF +NA G DQ+ LAR YI +K N+ + ++ +P ++
Sbjct 174 ATFLYPLLMASDIFAMNATHIIAGPDQKIHLTLAR----YIARKLNRHIDSNYFTVPQMV 229
Query 86 EGQDKMSKSNTDS-------------AIFMEDSEAEVNRKIKKA 116
+G KS ++ I M D++ ++ RKIK A
Sbjct 230 KGFTYKVKSLSNGTKKMSKSSESKFDTINMLDTDDDIYRKIKNA 273
> sce:YOL097C WRS1, HRE342; Wrs1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=432
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 46/97 (47%), Gaps = 10/97 (10%)
Query 50 GMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEV 109
+DQ + R+ D +K +K +L P L KMS S+ +AIFM D+ ++
Sbjct 256 AIDQDPYFRVCRDVADKLKY-SKPALLHSRFFPALQGSTTKMSASDDTTAIFMTDTPKQI 314
Query 110 NRKIKK-AFCPPCI-------IEGNPCISYA-EYLVY 137
+KI K AF + + GNP + A +YL +
Sbjct 315 QKKINKYAFSGGQVSADLHRELGGNPDVDVAYQYLSF 351
> mmu:640248 tryptophanyl-tRNA synthetase, cytoplasmic-like
Length=192
Score = 36.2 bits (82), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 12/100 (12%)
Query 27 AQIMYPCMQCA--------DIFYLNADI-CQL--GMDQRKVNVLAREYCDYIKKKNKQVI 75
+I +P +Q A IF DI C + +DQ + R+ I K +
Sbjct 48 GKISFPAVQAAPSFSNSFPKIFRDRTDIQCLIPCAIDQDPYFRMTRDVAPRIGHP-KPAL 106
Query 76 LSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
L P L Q KMS S+ +S+IF+ D+ ++ K+ K
Sbjct 107 LHSTFFPALQGAQTKMSASDPNSSIFLTDTAKQIKSKVNK 146
> eco:b3384 trpS, ECK3371, JW3347; tryptophanyl-tRNA synthetase
(EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=334
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 48/106 (45%), Gaps = 14/106 (13%)
Query 31 YPCMQCADIFYLNADICQLGMDQRKVNVLARE--------YCDYIKKKNKQVILSHHMLP 82
YP + ADI ++ +G DQ++ L+R+ Y + K + S +
Sbjct 128 YPVLMAADILLYQTNLVPVGEDQKQHLELSRDIAQRFNALYGEIFKVPEPFIPKSGARVM 187
Query 83 GLLEGQDKMSKS--NTDSAIFMEDSEAEVNRKIKKAFC----PPCI 122
LLE KMSKS N ++ I + + V +KIK+A PP +
Sbjct 188 SLLEPTKKMSKSDDNRNNVIGLLEDPKSVVKKIKRAVTDSDEPPVV 233
> xla:414492 wars, MGC81110, gamma-2, ifi53, ifp53; tryptophanyl-tRNA
synthetase (EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase
[EC:6.1.1.2]
Length=475
Score = 35.4 bits (80), Expect = 0.058, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 45/105 (42%), Gaps = 12/105 (11%)
Query 22 DDQPSAQIMYPCMQCA--------DIFYLNADICQL---GMDQRKVNVLAREYCDYIKKK 70
D +I +P +Q A +IF DI L +DQ + R+ I
Sbjct 276 DSDCIGKISFPAIQAAPSFSSSFPEIFKGRKDIQCLIPCAIDQDPYFRMTRDVAPRINYP 335
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
K ++ P L Q KMS S+ +S+IF+ D+ ++ KI K
Sbjct 336 -KPALMHSTFFPALQGAQTKMSASDPNSSIFLTDTAKQIKNKINK 379
> xla:380225 wars, MGC53671; tryptophanyl-tRNA synthetase (EC:6.1.1.2);
K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=475
Score = 35.4 bits (80), Expect = 0.064, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 45/105 (42%), Gaps = 12/105 (11%)
Query 22 DDQPSAQIMYPCMQCA--------DIFYLNADICQL---GMDQRKVNVLAREYCDYIKKK 70
D +I +P +Q A +IF DI L +DQ + R+ I
Sbjct 276 DSDCIGKISFPAIQAAPSFSSSFPEIFKGRKDIQCLIPCAIDQDPYFRMTRDVAPRINYP 335
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
K ++ P L Q KMS S+ +S+IF+ D+ ++ KI K
Sbjct 336 -KPALMHSTFFPALQGAQTKMSASDPNSSIFLTDTAKQIKSKINK 379
> cel:Y80D3A.1 wrs-1; tryptophanyl (W) tRNA Synthetase family
member (wrs-1); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=417
Score = 35.4 bits (80), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Query 50 GMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEV 109
+DQ + R+ +K +I S LP L Q KMS S ++ IF+ D+ ++
Sbjct 257 AIDQDPFFRMTRDVAPRLKASKPSLIFST-FLPALTGAQTKMSASEPNTCIFLSDTAKQI 315
Query 110 NRKIKK 115
KI K
Sbjct 316 KNKINK 321
> mmu:70560 Wars2, 5730427B17Rik, 9430020O07Rik, AI413375, TrpRS;
tryptophanyl tRNA synthetase 2 (mitochondrial) (EC:6.1.1.2);
K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=360
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 30/124 (24%), Positives = 54/124 (43%), Gaps = 11/124 (8%)
Query 6 ISRVKRCSQIMGRA--EGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREY 63
+ R++ Q +A + D + YP +Q ADI + +G DQ + L ++
Sbjct 133 LPRLQHLHQWKAKAAKQKHDGTVGLLTYPVLQAADILCYKSTHVPVGEDQVQHMELVQDL 192
Query 64 CDYIKKK-------NKQVILSHHMLPGLLEGQDKMSKSNTD--SAIFMEDSEAEVNRKIK 114
+K K ++ S + L + KMSKS+ D + + + DS E+ +K +
Sbjct 193 ARSFNQKYGEFFPLPKSILTSMKKVKSLRDPSSKMSKSDPDKLATVRITDSPEEIVQKFR 252
Query 115 KAFC 118
KA
Sbjct 253 KAVT 256
> dre:503745 wars2, zgc:110828; tryptophanyl tRNA synthetase 2,
mitochondrial (EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase
[EC:6.1.1.2]
Length=364
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 58/125 (46%), Gaps = 10/125 (8%)
Query 4 NSISRVKRCSQIMGRAEGDDQPSAQI-MYPCMQCADIFYLNADICQLGMDQRKVNVLARE 62
S+ R++ Q +++ ++ S + YP +Q ADI + +G DQ + LA++
Sbjct 136 TSMPRLRHLPQWKMKSKQKNEGSVGLYTYPVLQAADILLYKSTHVPVGEDQIQHLELAQD 195
Query 63 YCDYIKKKNKQVILS-HHMLPG------LLEGQDKMSKSNTD--SAIFMEDSEAEVNRKI 113
+ ++ +L G L + KMSKS+ SA+F+ D+ ++ KI
Sbjct 196 LARIFNNQYGEIFPEPRALLSGTRKVKSLRDPSSKMSKSDPQRFSAVFLNDTPDDIALKI 255
Query 114 KKAFC 118
++A
Sbjct 256 RRAVT 260
> mmu:22375 Wars, TrpRS, WRS; tryptophanyl-tRNA synthetase (EC:6.1.1.2);
K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=475
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 12/105 (11%)
Query 22 DDQPSAQIMYPCMQCA--------DIFYLNADICQL---GMDQRKVNVLAREYCDYIKKK 70
D +I +P +Q A IF DI L +DQ + R+ I
Sbjct 275 DSDCIGKISFPAVQAAPSFSNSFPKIFRDRTDIQCLIPCAIDQDPYFRMTRDVAPRIGHP 334
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
K +L P L Q KMS S+ +S+IF+ D+ ++ K+ K
Sbjct 335 -KPALLHSTFFPALQGAQTKMSASDPNSSIFLTDTAKQIKSKVNK 378
> hsa:10352 WARS2, TrpRS; tryptophanyl tRNA synthetase 2, mitochondrial
(EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase
[EC:6.1.1.2]
Length=360
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 45/99 (45%), Gaps = 9/99 (9%)
Query 29 IMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIKKK-------NKQVILSHHML 81
+ YP +Q ADI + +G DQ + L ++ KK + ++ S +
Sbjct 158 LTYPVLQAADILLYKSTHVPVGEDQVQHMELVQDLAQGFNKKYGEFFPVPESILTSMKKV 217
Query 82 PGLLEGQDKMSKSNTD--SAIFMEDSEAEVNRKIKKAFC 118
L + KMSKS+ D + + + DS E+ +K +KA
Sbjct 218 KSLRDPSAKMSKSDPDKLATVRITDSPEEIVQKFRKAVT 256
> hsa:7453 WARS, GAMMA-2, IFI53, IFP53; tryptophanyl-tRNA synthetase
(EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=471
Score = 34.3 bits (77), Expect = 0.15, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 12/105 (11%)
Query 22 DDQPSAQIMYPCMQCA--------DIFYLNADICQL---GMDQRKVNVLAREYCDYIKKK 70
D +I +P +Q A IF DI L +DQ + R+ I
Sbjct 271 DSDCIGKISFPAIQAAPSFSNSFPQIFRDRTDIQCLIPCAIDQDPYFRMTRDVAPRIGYP 330
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
K +L P L Q KMS S+ +S+IF+ D+ ++ K+ K
Sbjct 331 -KPALLHSTFFPALQGAQTKMSASDPNSSIFLTDTAKQIKTKVNK 374
> dre:393745 wars, MGC73274, zgc:73274; tryptophanyl-tRNA synthetase
(EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=463
Score = 33.9 bits (76), Expect = 0.17, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 72 KQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
K +L P L Q KMS S+ +S IF+ D+ ++ K+ K
Sbjct 324 KPALLHSTFFPALQGAQTKMSASDANSTIFLTDTPKQIKNKVNK 367
> tpv:TP01_0518 tryptophanyl-tRNA synthetase; K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=579
Score = 32.7 bits (73), Expect = 0.39, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 42/98 (42%), Gaps = 12/98 (12%)
Query 29 IMYPCMQCA--------DIFYLNADICQL---GMDQRKVNVLAREYCDYIKKKNKQVILS 77
I YP ++ A ++F DI L G+DQ L R + K K ++
Sbjct 384 ISYPLLEGAASFSESFPNLFNNKEDIMCLVPQGIDQDPFFRLTRSLAPKLGLK-KPALIH 442
Query 78 HHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
+P LL KMS S SAIF+ D + +KI K
Sbjct 443 SKFIPSLLGINQKMSSSIEGSAIFVTDDAETIKKKIHK 480
> pfa:PFL2485c tryptophanyl-tRNA synthetase, putative (EC:6.1.1.2);
K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=570
Score = 32.3 bits (72), Expect = 0.54, Method: Composition-based stats.
Identities = 30/118 (25%), Positives = 50/118 (42%), Gaps = 14/118 (11%)
Query 13 SQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREYCDYIK---- 68
S I + G ++ A YP + ADI +G DQ K + ++ C I
Sbjct 266 SHIHIKMGGHNKSVALFSYPSLMLADILLYKPTYLIIGQDQIKNMEIMKKLCRKINYHFS 325
Query 69 --KKNKQVILSHHMLPGL-LEGQDKMSKS-------NTDSAIFMEDSEAEVNRKIKKA 116
K ++ S + L+G KMSK+ +T I++ D++ + KIKK+
Sbjct 326 NVAKLPKIFFSKFYSEVMNLDGHKKMSKNHLLHHDKDTSKIIYLLDNKKTIENKIKKS 383
> bbo:BBOV_IV005840 23.m06426; serine hydroxymethyltransferase
(EC:2.1.2.1); K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=453
Score = 32.3 bits (72), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 32/70 (45%), Gaps = 17/70 (24%)
Query 79 HMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNR-----------KIKKAFCPPCIIEGNP 127
H+ G GQ K+S + A+F + +VN+ ++ KA+CP II G
Sbjct 138 HLTHGFYVGQKKISAT----AVFYTSLQYDVNKETGLLDYDDMERLAKAYCPKLIIAGAS 193
Query 128 CISYAEYLVY 137
C Y+ Y Y
Sbjct 194 C--YSRYWDY 201
> cpv:cgd7_1490 tryptophanyl-tRNA synthetase ; K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=593
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 80 MLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
LP L Q KMS S +S+IF+ D+E + KI K
Sbjct 456 FLPSLQGSQTKMSASVQNSSIFVNDNEESIRNKIMK 491
> ath:AT2G25840 OVA4; OVA4 (ovule abortion 4); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / tryptophan-tRNA ligase
(EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=408
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 18 RAEGDDQPSAQIM-YPCMQCADIFYLNADICQLGMDQRKVNVLARE 62
R EG + S + YP + ADI +D +G DQ++ LARE
Sbjct 176 RKEGVENASVGLFTYPDLMTADILLYQSDFVPVGEDQKQHIELARE 221
> cpv:cgd1_3170 hypothetical protein
Length=1619
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 6/45 (13%)
Query 42 LNADICQLGMDQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLE 86
+N D C + D+RK+++L IK KN+QV ++M P L E
Sbjct 709 INVDECTIECDRRKMSIL------LIKVKNRQVESFYYMFPSLEE 747
> dre:558044 spectrin, beta, non-erythrocytic 1; K06115 spectrin
beta
Length=2391
Score = 29.6 bits (65), Expect = 3.2, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVN 110
+++ +LSH +P L+G ++ K + D ME SE ++N
Sbjct 1226 SQEYVLSHTEMPSSLQGAEEAIKKHEDFLTTMEASEEKIN 1265
> ath:AT3G04600 tRNA synthetase class I (W and Y) family protein
(EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=402
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
+K ++ P L KMS S+ +SAI++ DS ++ KI +
Sbjct 261 SKPALIESTFFPALQGENGKMSASDPNSAIYVTDSAKDIKNKINR 305
> hsa:9779 TBC1D5, KIAA0210; TBC1 domain family, member 5
Length=795
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 26/91 (28%), Positives = 48/91 (52%), Gaps = 5/91 (5%)
Query 4 NSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREY 63
++ISR + S+ MGR E +++ AQI + Q D+ + C MD VN+
Sbjct 567 SNISRSRSHSKTMGRKESEEELEAQISFLQGQLNDLDAM-CKYCAKVMDTHLVNIQDVIL 625
Query 64 CDYIKKKNKQVILSHHMLPGLLEGQDKMSKS 94
+ ++K++ Q+++S L GL + +D + S
Sbjct 626 QENLEKED-QILVS---LAGLKQIKDILKGS 652
> mmu:229589 Prune, 9230112O05Rik, C130058A12, DRES-17, HTCD37,
PRUNEM1, Prune-M1; prune homolog (Drosophila) (EC:3.6.1.1);
K01514 exopolyphosphatase [EC:3.6.1.11]
Length=454
Score = 28.9 bits (63), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 29/50 (58%), Gaps = 16/50 (32%)
Query 74 VILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEV--NRKIKKAFCPPC 121
+++ HH+LP +D+A +E++ AEV +R I++ +CPPC
Sbjct 103 ILVDHHILP------------KSDAA--LEEAVAEVLDHRPIEQKYCPPC 138
> mmu:72238 Tbc1d5, 1600014N05Rik, KIAA0210, mKIAA0210; TBC1 domain
family, member 5
Length=815
Score = 28.9 bits (63), Expect = 6.6, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 48/91 (52%), Gaps = 5/91 (5%)
Query 4 NSISRVKRCSQIMGRAEGDDQPSAQIMYPCMQCADIFYLNADICQLGMDQRKVNVLAREY 63
++I+R + S+ MGR E +++ AQI + Q D+ + C MD VN+
Sbjct 591 SNIARSRSHSKTMGRKESEEELEAQISFLQGQLNDLDAM-CKYCAKVMDMHLVNIQDVVL 649
Query 64 CDYIKKKNKQVILSHHMLPGLLEGQDKMSKS 94
+ ++K++ Q+++S L GL + +D + S
Sbjct 650 QENLEKED-QILVS---LAGLKQIKDILKGS 676
> cpv:cgd2_1260 hypothetical protein
Length=811
Score = 28.5 bits (62), Expect = 6.9, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 27/62 (43%), Gaps = 0/62 (0%)
Query 52 DQRKVNVLAREYCDYIKKKNKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNR 111
D+ N+L Y DY + L LP + + Q+K+ + D +E E+N
Sbjct 46 DESITNILNAAYSDYSLISTDLLPLVDSYLPSIKQFQEKIDSNVNDVTSIVERYSEEINS 105
Query 112 KI 113
KI
Sbjct 106 KI 107
> hsa:6711 SPTBN1, ELF, SPTB2, betaSpII; spectrin, beta, non-erythrocytic
1; K06115 spectrin beta
Length=2364
Score = 28.5 bits (62), Expect = 8.6, Method: Composition-based stats.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVN 110
N++ +L+H +P LEG + K D M+ +E ++N
Sbjct 1187 NQEYVLAHTEMPTTLEGAEAAIKKQEDFMTTMDANEEKIN 1226
> mmu:20742 Spnb2, 9930031C03Rik, AL033301, KIAA4049, SPTB2, SPTBN1,
Spnb-2, elf1, elf3, mKIAA4049; spectrin beta 2; K06115
spectrin beta
Length=2154
Score = 28.5 bits (62), Expect = 8.7, Method: Composition-based stats.
Identities = 12/40 (30%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 71 NKQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVN 110
N++ +L+H +P LEG + K D M+ +E ++N
Sbjct 1174 NQEYVLAHTEMPTTLEGAEAAIKKQEDFMTTMDANEEKIN 1213
> bbo:BBOV_III002530 17.m07244; tryptophanyl-tRNA synthetase family
protein (EC:6.1.1.2); K01867 tryptophanyl-tRNA synthetase
[EC:6.1.1.2]
Length=587
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 72 KQVILSHHMLPGLLEGQDKMSKSNTDSAIFMEDSEAEVNRKIKK 115
K + + +P LL KMS S SAIF+ D+ + K+ K
Sbjct 444 KPISIHSKFIPSLLGVTQKMSSSIEGSAIFVTDTPKMIRDKVHK 487
Lambda K H
0.321 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40