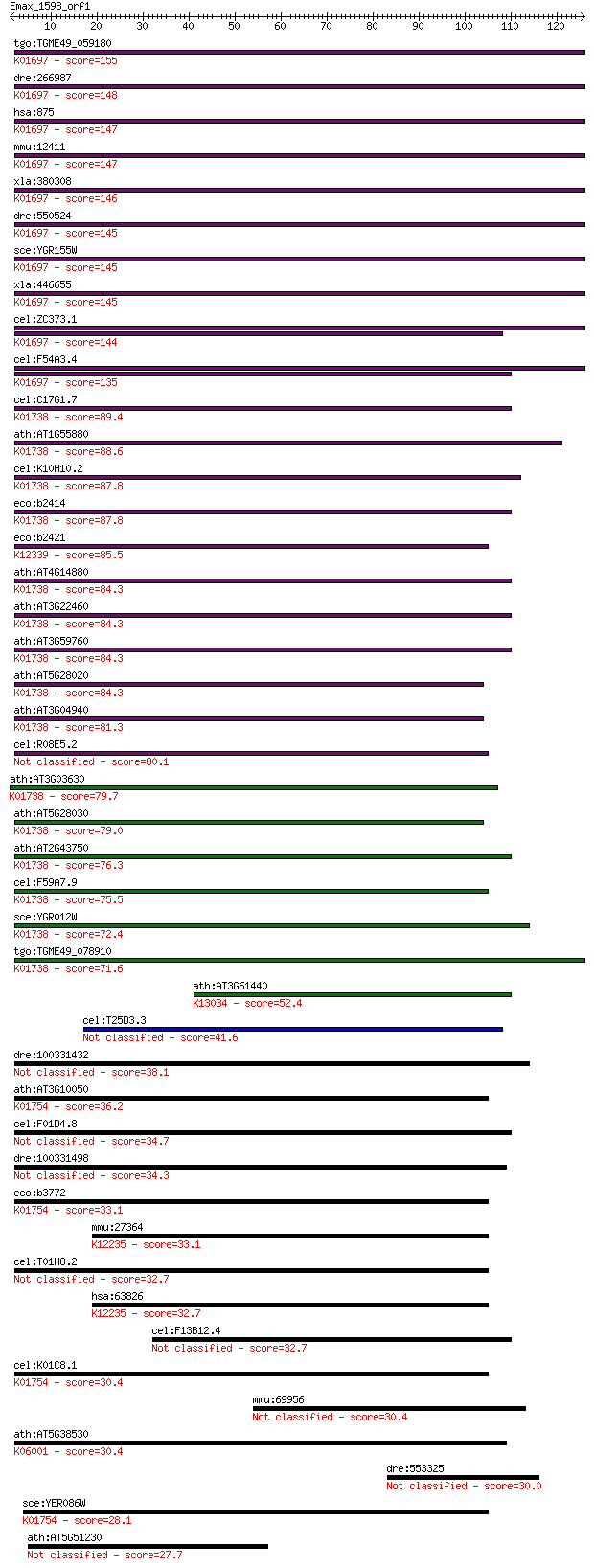
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1598_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_059180 cystathionine beta-synthase, putative (EC:4.... 155 2e-38
dre:266987 cbsb, cb442, cbs, wu:fb37g05, wu:fm61c07, wu:fq06c0... 148 3e-36
hsa:875 CBS, HIP4; cystathionine-beta-synthase (EC:4.2.1.22); ... 147 7e-36
mmu:12411 Cbs, AI047524, AI303044, HIP4, MGC18856, MGC18895, M... 147 8e-36
xla:380308 cbs-a, MGC52737, hip4; cystathionine-beta-synthase ... 146 2e-35
dre:550524 cbsa, MGC174292, zgc:110199; cystathionine-beta-syn... 145 2e-35
sce:YGR155W CYS4, NHS5, STR4, VMA41; Cys4p (EC:4.2.1.22); K016... 145 2e-35
xla:446655 cbs-b, MGC82640, cbs, hip4; cystathionine-beta-synt... 145 4e-35
cel:ZC373.1 hypothetical protein; K01697 cystathionine beta-sy... 144 8e-35
cel:F54A3.4 hypothetical protein; K01697 cystathionine beta-sy... 135 3e-32
cel:C17G1.7 hypothetical protein; K01738 cysteine synthase A [... 89.4 2e-18
ath:AT1G55880 pyridoxal-5'-phosphate-dependent enzyme, beta fa... 88.6 4e-18
cel:K10H10.2 hypothetical protein; K01738 cysteine synthase A ... 87.8 7e-18
eco:b2414 cysK, cysZ, ECK2409, JW2407; cysteine synthase A, O-... 87.8 7e-18
eco:b2421 cysM, ECK2416, JW2414; cysteine synthase B (O-acetyl... 85.5 4e-17
ath:AT4G14880 OASA1; OASA1 (O-ACETYLSERINE (THIOL) LYASE (OAS-... 84.3 8e-17
ath:AT3G22460 OASA2; OASA2 (O-ACETYLSERINE (THIOL) LYASE (OAS-... 84.3 9e-17
ath:AT3G59760 OASC; OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM... 84.3 9e-17
ath:AT5G28020 CYSD2; CYSD2 (CYSTEINE SYNTHASE D2); catalytic/ ... 84.3 1e-16
ath:AT3G04940 CYSD1; CYSD1 (CYSTEINE SYNTHASE D1); cysteine sy... 81.3 7e-16
cel:R08E5.2 hypothetical protein 80.1 1e-15
ath:AT3G03630 CS26; CS26; cysteine synthase; K01738 cysteine s... 79.7 2e-15
ath:AT5G28030 cysteine synthase, putative / O-acetylserine (th... 79.0 4e-15
ath:AT2G43750 OASB; OASB (O-ACETYLSERINE (THIOL) LYASE B); cys... 76.3 2e-14
cel:F59A7.9 hypothetical protein; K01738 cysteine synthase A [... 75.5 4e-14
sce:YGR012W Putative cysteine synthase, localized to the mitoc... 72.4 3e-13
tgo:TGME49_078910 O-acetylserine (thiol) lyase, putative (EC:2... 71.6 6e-13
ath:AT3G61440 CYSC1; CYSC1 (CYSTEINE SYNTHASE C1); L-3-cyanoal... 52.4 3e-07
cel:T25D3.3 hypothetical protein 41.6 6e-04
dre:100331432 serine racemase-like 38.1 0.007
ath:AT3G10050 OMR1; OMR1 (L-O-METHYLTHREONINE RESISTANT 1); L-... 36.2 0.027
cel:F01D4.8 hypothetical protein 34.7 0.073
dre:100331498 serine racemase-like 34.3 0.11
eco:b3772 ilvA, ECK3764, JW3745; threonine deaminase (EC:4.3.1... 33.1 0.21
mmu:27364 Srr, Srs; serine racemase (EC:4.3.1.17 5.1.1.18 4.3.... 33.1 0.21
cel:T01H8.2 hypothetical protein 32.7 0.27
hsa:63826 SRR, ILV1, ISO1; serine racemase (EC:4.3.1.17 5.1.1.... 32.7 0.28
cel:F13B12.4 hypothetical protein 32.7 0.32
cel:K01C8.1 hypothetical protein; K01754 threonine dehydratase... 30.4 1.3
mmu:69956 Ptcd3, 2610034F17Rik, 2810422B04Rik, AA589622, AU045... 30.4 1.3
ath:AT5G38530 tryptophan synthase-related; K06001 tryptophan s... 30.4 1.5
dre:553325 ptcd3, MGC123014, fa08d08, wu:fa08d08, zgc:123014; ... 30.0 1.7
sce:YER086W ILV1, ISO1; Ilv1p (EC:4.3.1.19); K01754 threonine ... 28.1 7.9
ath:AT5G51230 EMF2; EMF2 (EMBRYONIC FLOWER 2); DNA binding / t... 27.7 8.5
> tgo:TGME49_059180 cystathionine beta-synthase, putative (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=514
Score = 155 bits (393), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 71/124 (57%), Positives = 90/124 (72%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TP+VR+ R+A G+ C LL KCEF+S GGS KDRI + M+ AE G L G LIEPT
Sbjct 24 TPMVRMKRLAKVYGLECDLLAKCEFMSAGGSVKDRIGKAMVEKAEREGRLKAGDTLIEPT 83
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + MP KMS EK IMK LG+ I+RTPT AAW+++ SH+ +
Sbjct 84 SGNTGIGLALAAAVRGYRMIVTMPAKMSAEKSNIMKCLGAEIVRTPTEAAWNDENSHMGV 143
Query 122 SLRL 125
+ +L
Sbjct 144 AAKL 147
> dre:266987 cbsb, cb442, cbs, wu:fb37g05, wu:fm61c07, wu:fq06c06,
zgc:113704; cystathionine-beta-synthase b (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=597
Score = 148 bits (374), Expect = 3e-36, Method: Composition-based stats.
Identities = 69/124 (55%), Positives = 92/124 (74%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TP+VR++++ A G+ C+LL KCEF + GGS KDRI+ MI AE G L PG +IEPT
Sbjct 122 TPMVRMNKIPKAFGLKCELLAKCEFFNAGGSVKDRISLRMIEDAERDGTLKPGDTIIEPT 181
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + VMP KMS EK +++ALG+ I+RTPT+A +D SH+ +
Sbjct 182 SGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTSARFDSPESHVGV 241
Query 122 SLRL 125
+ RL
Sbjct 242 AWRL 245
> hsa:875 CBS, HIP4; cystathionine-beta-synthase (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=551
Score = 147 bits (371), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 68/124 (54%), Positives = 91/124 (73%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TP+VR++++ G+ C+LL KCEF + GGS KDRI+ MI AE G L PG +IEPT
Sbjct 87 TPMVRINKIGKKFGLKCELLAKCEFFNAGGSVKDRISLRMIEDAERDGTLKPGDTIIEPT 146
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + VMP KMS+EK +++ALG+ I+RTPT A +D SH+ +
Sbjct 147 SGNTGIGLALAAAVRGYRCIIVMPEKMSSEKVDVLRALGAEIVRTPTNARFDSPESHVGV 206
Query 122 SLRL 125
+ RL
Sbjct 207 AWRL 210
> mmu:12411 Cbs, AI047524, AI303044, HIP4, MGC18856, MGC18895,
MGC37300; cystathionine beta-synthase (EC:4.2.1.22); K01697
cystathionine beta-synthase [EC:4.2.1.22]
Length=561
Score = 147 bits (371), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 68/124 (54%), Positives = 93/124 (75%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TP+VR+++++ G+ C+LL KCEF + GGS KDRI+ MI AE +G+L PG +IEPT
Sbjct 84 TPMVRINKISKNAGLKCELLAKCEFFNAGGSVKDRISLRMIEDAERAGNLKPGDTIIEPT 143
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + VMP KMS EK +++ALG+ I+RTPT A +D SH+ +
Sbjct 144 SGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTNARFDSPESHVGV 203
Query 122 SLRL 125
+ RL
Sbjct 204 AWRL 207
> xla:380308 cbs-a, MGC52737, hip4; cystathionine-beta-synthase
(EC:4.2.1.22); K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=565
Score = 146 bits (369), Expect = 2e-35, Method: Composition-based stats.
Identities = 68/124 (54%), Positives = 91/124 (73%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPLVR++++ G+ C+LL KCEF + GGS KDRI+ M+ AE G L PG +IEPT
Sbjct 83 TPLVRINQIGKRFGLKCELLAKCEFFNAGGSVKDRISLRMVEDAERDGILKPGDTIIEPT 142
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + VMP KMS EK +++ALG+ I+RTPT+A +D SH+ +
Sbjct 143 SGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTSARFDSPESHVGV 202
Query 122 SLRL 125
+ RL
Sbjct 203 AWRL 206
> dre:550524 cbsa, MGC174292, zgc:110199; cystathionine-beta-synthase
a; K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=571
Score = 145 bits (367), Expect = 2e-35, Method: Composition-based stats.
Identities = 67/124 (54%), Positives = 91/124 (73%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPLV +++++ G+ C+LL KCEF + GGS KDRI+ M+ AE G L PG +IEPT
Sbjct 98 TPLVHINKISKIFGLKCELLAKCEFFNAGGSVKDRISLRMVEDAEREGILNPGDTIIEPT 157
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + VMP KMS EK +++ALG+ I+RTPT+A +D SH+ +
Sbjct 158 SGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTSARFDSPESHVGV 217
Query 122 SLRL 125
+ RL
Sbjct 218 AWRL 221
> sce:YGR155W CYS4, NHS5, STR4, VMA41; Cys4p (EC:4.2.1.22); K01697
cystathionine beta-synthase [EC:4.2.1.22]
Length=507
Score = 145 bits (367), Expect = 2e-35, Method: Composition-based stats.
Identities = 69/125 (55%), Positives = 89/125 (71%), Gaps = 1/125 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TPL+ L ++ ALGI Q+ K E +PGGS KDRIA+ M+ AEASG + P LIEP
Sbjct 21 TPLIALKKLPKALGIKPQIYAKLELYNPGGSIKDRIAKSMVEEAEASGRIHPSRSTLIEP 80
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIA 120
TSGNTGIGLA+ A+ GYR++ +P KMS EK ++KALG+ I+RTPTAAAWD SHI
Sbjct 81 TSGNTGIGLALIGAIKGYRTIITLPEKMSNEKVSVLKALGAEIIRTPTAAAWDSPESHIG 140
Query 121 LSLRL 125
++ +L
Sbjct 141 VAKKL 145
> xla:446655 cbs-b, MGC82640, cbs, hip4; cystathionine-beta-synthase
(EC:4.2.1.22); K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=562
Score = 145 bits (365), Expect = 4e-35, Method: Composition-based stats.
Identities = 68/124 (54%), Positives = 91/124 (73%), Gaps = 0/124 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPLVR++++ G+ C+LL KCEF + GGS KDRI+ M+ AE G L PG +IEPT
Sbjct 80 TPLVRINQIGRRFGLKCELLAKCEFFNAGGSVKDRISLRMVEDAERDGILKPGDTIIEPT 139
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIAL 121
SGNTGIGLA+A+AV GYR + VMP KMS EK +++ALG+ I+RTPT+A +D SH+ +
Sbjct 140 SGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTSARFDSPESHVGV 199
Query 122 SLRL 125
+ RL
Sbjct 200 AWRL 203
> cel:ZC373.1 hypothetical protein; K01697 cystathionine beta-synthase
[EC:4.2.1.22]
Length=704
Score = 144 bits (362), Expect = 8e-35, Method: Composition-based stats.
Identities = 70/127 (55%), Positives = 89/127 (70%), Gaps = 3/127 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGH---LAPGCALI 58
TPLV+L + A G+ C + KCE+++ GGSTKDRIA+ M+ AE +G L PG LI
Sbjct 389 TPLVKLQHIPKAHGVKCNVYVKCEYMNAGGSTKDRIAKRMVEIAEKTGKPGKLVPGVTLI 448
Query 59 EPTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSH 118
EPTSGNTGIGL++ASAV GY+ + MP KMS EK M +LGSTI+RTP A +D SH
Sbjct 449 EPTSGNTGIGLSLASAVRGYKCIITMPKKMSKEKSIAMASLGSTIIRTPNEAGFDSPHSH 508
Query 119 IALSLRL 125
I ++LRL
Sbjct 509 IGVALRL 515
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 27/106 (25%), Positives = 43/106 (40%), Gaps = 1/106 (0%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPLV L ++ + + K E+L+ GS +DR A + AE G + G +
Sbjct 30 TPLVPLKKLKVEHQLQSDIYVKLEYLNIAGSLEDRTADKAFQFAEEIG-VVRGDEVFVTA 88
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTP 107
G+ I A +AV G + P ++ LG + P
Sbjct 89 GGSAAISYATVAAVKGIKLTIYAPKGEFDLVDTVLHTLGVKTVELP 134
> cel:F54A3.4 hypothetical protein; K01697 cystathionine beta-synthase
[EC:4.2.1.22]
Length=755
Score = 135 bits (341), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 70/128 (54%), Positives = 89/128 (69%), Gaps = 4/128 (3%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGH---LAPGCA-L 57
TPLV+L V A G+ C + KCEFL+ GGSTKDRIA+ M+ AE +G L PG L
Sbjct 347 TPLVKLQHVPKAHGVRCNVYVKCEFLNAGGSTKDRIAKKMVEIAEKTGKPGALTPGATTL 406
Query 58 IEPTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTS 117
IEPTSGNTGIGL++ +AV GY+ + MP KMS EK + LGSTI+RTP AA++ +S
Sbjct 407 IEPTSGNTGIGLSLVAAVRGYKCLITMPEKMSKEKSTTLSVLGSTIVRTPNEAAFNSPSS 466
Query 118 HIALSLRL 125
HI ++LRL
Sbjct 467 HIGVALRL 474
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 49/108 (45%), Gaps = 3/108 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
T LV L ++ A + + K E+L+ GS +DR A RA E S + G +
Sbjct 21 TQLVPLKKLKAEFQLKSDIYVKLEYLNISGSLEDRAAH---RAFEMSNGIQRGDEVYVTA 77
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
G++ + A +AV G R A P + I+ LG+ I+ P +
Sbjct 78 GGSSAVSYATVAAVKGVRLTAHAPPGAFDQVRTILSTLGTKIVELPVS 125
> cel:C17G1.7 hypothetical protein; K01738 cysteine synthase A
[EC:2.5.1.47]
Length=341
Score = 89.4 bits (220), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/109 (44%), Positives = 67/109 (61%), Gaps = 3/109 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCA-LIEP 60
TP+V ++++ G+ + K E+++P GS KDRI M+ AAE G + PG LIEP
Sbjct 18 TPMVYINKLTK--GLPGTVAVKIEYMNPAGSVKDRIGAAMLAAAEKDGTVIPGVTTLIEP 75
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
TSGNTGI LA +A GYR + MP MS E+ ++KA GS ++ T A
Sbjct 76 TSGNTGIALAFVAAAKGYRCIVTMPASMSGERRTLLKAYGSEVVLTDPA 124
> ath:AT1G55880 pyridoxal-5'-phosphate-dependent enzyme, beta
family protein (EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=421
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 76/120 (63%), Gaps = 3/120 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPL+R++ ++ A G C++LGKCEFL+PGGS KDR+A +I+ A SG L PG + E +
Sbjct 51 TPLIRINSLSEATG--CEILGKCEFLNPGGSVKDRVAVKIIQEALESGKLFPGGIVTEGS 108
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRT-PTAAAWDEQTSHIA 120
+G+T I LA + G + V+P + EK +I++ALG+++ R P + + +IA
Sbjct 109 AGSTAISLATVAPAYGCKCHVVIPDDAAIEKSQIIEALGASVERVRPVSITHKDHYVNIA 168
> cel:K10H10.2 hypothetical protein; K01738 cysteine synthase
A [EC:2.5.1.47]
Length=337
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/111 (43%), Positives = 68/111 (61%), Gaps = 3/111 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TPL++L+++ LG S + K E+++P S KDRIA MI AE +G + PG LIEP
Sbjct 17 TPLLKLNKIGKDLGASIAV--KVEYMNPACSVKDRIAFNMIDTAEKAGLITPGKTVLIEP 74
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAA 111
TSGN GI LA + GY+ + MP MS E+ ++KA G+ ++ T A A
Sbjct 75 TSGNMGIALAYCGKLRGYKVILTMPASMSIERRCLLKAYGAEVILTDPATA 125
> eco:b2414 cysK, cysZ, ECK2409, JW2407; cysteine synthase A,
O-acetylserine sulfhydrolase A subunit (EC:2.5.1.47); K01738
cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 49/108 (45%), Positives = 65/108 (60%), Gaps = 5/108 (4%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPLVRL+R+ + ++L K E +P S K RI MI AE G L PG L+EPT
Sbjct 15 TPLVRLNRIG-----NGRILAKVESRNPSFSVKCRIGANMIWDAEKRGVLKPGVELVEPT 69
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
SGNTGI LA +A GY+ MP MS E+ +++KALG+ ++ T A
Sbjct 70 SGNTGIALAYVAAARGYKLTLTMPETMSIERRKLLKALGANLVLTEGA 117
> eco:b2421 cysM, ECK2416, JW2414; cysteine synthase B (O-acetylserine
sulfhydrolase B) (EC:2.5.1.47); K12339 cysteine synthase
B [EC:2.5.1.47]
Length=303
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 49/103 (47%), Positives = 61/103 (59%), Gaps = 2/103 (1%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPLV+L R+ G L K E +P GS KDR A MI AE G + PG LIE T
Sbjct 11 TPLVKLQRMGPDNGSEVWL--KLEGNNPAGSVKDRAALSMIVEAEKRGEIKPGDVLIEAT 68
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
SGNTGI LAM +A+ GYR +MP MS E+ M+A G+ ++
Sbjct 69 SGNTGIALAMIAALKGYRMKLLMPDNMSQERRAAMRAYGAELI 111
> ath:AT4G14880 OASA1; OASA1 (O-ACETYLSERINE (THIOL) LYASE (OAS-TL)
ISOFORM A1); cysteine synthase (EC:2.5.1.47); K01738 cysteine
synthase A [EC:2.5.1.47]
Length=322
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 50/109 (45%), Positives = 63/109 (57%), Gaps = 3/109 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TPLV L+ VA G ++ K E + P S KDRI MI AE G + PG LIEP
Sbjct 16 TPLVYLNNVAE--GCVGRVAAKLEMMEPCSSVKDRIGFSMISDAEKKGLIKPGESVLIEP 73
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
TSGNTG+GLA +A GY+ + MP MSTE+ I+ A G ++ T A
Sbjct 74 TSGNTGVGLAFTAAAKGYKLIITMPASMSTERRIILLAFGVELVLTDPA 122
> ath:AT3G22460 OASA2; OASA2 (O-ACETYLSERINE (THIOL) LYASE (OAS-TL)
ISOFORM A1); catalytic/ cysteine synthase/ pyridoxal phosphate
binding; K01738 cysteine synthase A [EC:2.5.1.47]
Length=188
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 51/111 (45%), Positives = 64/111 (57%), Gaps = 7/111 (6%)
Query 2 TPLVRLSRVAAALGISC--QLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALI 58
TPLV L++VA C + K E + P S KDRI MI AEA G + PG LI
Sbjct 19 TPLVYLNKVAK----DCVGHVAAKLEMMEPCSSVKDRIGYSMIADAEAKGLIKPGESVLI 74
Query 59 EPTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
EPTSGNTG+GLA +A GY+ V MP MS E+ I+ A G+ ++ T A
Sbjct 75 EPTSGNTGVGLAFTAAAKGYKLVITMPASMSIERRIILLAFGAELILTDPA 125
> ath:AT3G59760 OASC; OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM
C); ATP binding / cysteine synthase (EC:2.5.1.47); K01738
cysteine synthase A [EC:2.5.1.47]
Length=430
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 47/109 (43%), Positives = 64/109 (58%), Gaps = 3/109 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TP+V L+ +A G + K E + P S KDRI M+ AE G ++PG L+EP
Sbjct 124 TPMVYLNSIAK--GCVANIAAKLEIMEPCCSVKDRIGYSMVTDAEQKGFISPGKSVLVEP 181
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
TSGNTGIGLA +A GYR + MP MS E+ ++KA G+ ++ T A
Sbjct 182 TSGNTGIGLAFIAASRGYRLILTMPASMSMERRVLLKAFGAELVLTDPA 230
> ath:AT5G28020 CYSD2; CYSD2 (CYSTEINE SYNTHASE D2); catalytic/
cysteine synthase/ pyridoxal phosphate binding (EC:2.5.1.47);
K01738 cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 48/103 (46%), Positives = 63/103 (61%), Gaps = 3/103 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TP+V L+ V G ++ K E + P S KDRIA MI+ AE G + PG LIEP
Sbjct 18 TPMVYLNNVVD--GCVARIAAKLEMMEPCSSVKDRIAYSMIKDAEDKGLITPGKSTLIEP 75
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTI 103
T+GNTGIGLA A GY+ + VMP MS E+ I++ALG+ +
Sbjct 76 TAGNTGIGLACMGAARGYKVILVMPSTMSLERRIILRALGAEL 118
> ath:AT3G04940 CYSD1; CYSD1 (CYSTEINE SYNTHASE D1); cysteine
synthase (EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=324
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 47/103 (45%), Positives = 61/103 (59%), Gaps = 3/103 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TP+V L+ + G ++ K E + P S K+RIA GMI+ AE G + PG LIE
Sbjct 19 TPMVYLNNIVD--GCVARIAAKLEMMEPCSSVKERIAYGMIKDAEDKGLITPGKSTLIEA 76
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTI 103
TSGNTGIGLA A GY+ V MP MS E+ I+ ALG+ +
Sbjct 77 TSGNTGIGLAFIGAAKGYKVVLTMPSSMSLERKIILLALGAEV 119
> cel:R08E5.2 hypothetical protein
Length=337
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 42/104 (40%), Positives = 64/104 (61%), Gaps = 3/104 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TPLV L ++ G+ ++ K E+L+P S KDRIA+ M+ AE +G + PG L+E
Sbjct 17 TPLVLLRNISK--GLDARIAVKVEYLNPSCSVKDRIAKSMVDEAEKAGTIVPGKTVLVEG 74
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
TSGN GI LA + GY+ + VMP MS E+ +++A G+ ++
Sbjct 75 TSGNLGIALAHIGKIRGYKVILVMPATMSVERRAMLRAYGAEVI 118
> ath:AT3G03630 CS26; CS26; cysteine synthase; K01738 cysteine
synthase A [EC:2.5.1.47]
Length=404
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/107 (40%), Positives = 63/107 (58%), Gaps = 3/107 (2%)
Query 1 CTPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAP-GCALIE 59
TP+V L+RV G + K E + P S KDRI MI AE SG + P L+E
Sbjct 109 STPMVYLNRVTD--GCLADIAAKLESMEPCRSVKDRIGLSMINEAENSGAITPRKTVLVE 166
Query 60 PTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRT 106
PT+GNTG+G+A +A GY+ + MP ++ E+ +++ALG+ I+ T
Sbjct 167 PTTGNTGLGIAFVAAAKGYKLIVTMPASINIERRMLLRALGAEIVLT 213
> ath:AT5G28030 cysteine synthase, putative / O-acetylserine (thiol)-lyase,
putative / O-acetylserine sulfhydrylase, putative
(EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 45/103 (43%), Positives = 62/103 (60%), Gaps = 3/103 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TP+V L+++ G ++ K E + P S KDRIA MI+ AE G + PG LIE
Sbjct 18 TPMVYLNKIVD--GCVARIAAKLEMMEPCSSIKDRIAYSMIKDAEDKGLITPGKSTLIEA 75
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTI 103
T GNTGIGLA A GY+ + +MP MS E+ I++ALG+ +
Sbjct 76 TGGNTGIGLASIGASRGYKVILLMPSTMSLERRIILRALGAEV 118
> ath:AT2G43750 OASB; OASB (O-ACETYLSERINE (THIOL) LYASE B); cysteine
synthase; K01738 cysteine synthase A [EC:2.5.1.47]
Length=392
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/109 (41%), Positives = 61/109 (55%), Gaps = 3/109 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TP+V L+ V G + K E + P S KDRI MI AE G + PG L+E
Sbjct 86 TPMVYLNNVVK--GCVASVAAKLEIMEPCCSVKDRIGYSMITDAEEKGLITPGKSVLVES 143
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
TSGNTGIGLA +A GY+ + MP MS E+ +++A G+ ++ T A
Sbjct 144 TSGNTGIGLAFIAASKGYKLILTMPASMSLERRVLLRAFGAELVLTEPA 192
> cel:F59A7.9 hypothetical protein; K01738 cysteine synthase A
[EC:2.5.1.47]
Length=337
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 62/104 (59%), Gaps = 3/104 (2%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPG-CALIEP 60
TPLV L + G+ ++ K E+L+P S KDRIA+ M+ AE +G + PG L+E
Sbjct 17 TPLVLLRNITK--GLDARIAVKIEYLNPSCSVKDRIAKTMVDDAEKAGKIVPGKTVLVEG 74
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
TSGN GI LA + GY+ + VM MS E+ +++A G+ ++
Sbjct 75 TSGNLGIALAHIGRIRGYKVILVMTTAMSIERRAMLRAYGAEVI 118
> sce:YGR012W Putative cysteine synthase, localized to the mitochondrial
outer membrane (EC:2.5.1.47); K01738 cysteine synthase
A [EC:2.5.1.47]
Length=393
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 41/114 (35%), Positives = 66/114 (57%), Gaps = 4/114 (3%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCA--LIE 59
TPLV++ + A G++ + K E +P GS KDR+A +I+ AE G L G + E
Sbjct 56 TPLVKIRSLTKATGVN--IYAKLELCNPAGSAKDRVALNIIKTAEELGELVRGEPGWVFE 113
Query 60 PTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWD 113
TSG+TGI +A+ +GYR+ +P S EK ++++LG+T+ + A+ D
Sbjct 114 GTSGSTGISIAVVCNALGYRAHISLPDDTSLEKLALLESLGATVNKVKPASIVD 167
> tgo:TGME49_078910 O-acetylserine (thiol) lyase, putative (EC:2.5.1.47);
K01738 cysteine synthase A [EC:2.5.1.47]
Length=499
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 46/125 (36%), Positives = 63/125 (50%), Gaps = 15/125 (12%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHL-APGCALIEP 60
TPL+ L R A+ C + KCEF +PGGS KDR A+ ++ AE G L ++E
Sbjct 155 TPLIYLRR--ASQETHCDIFAKCEFNNPGGSVKDRPAKYILEQAEKEGRLGGSQNWIVEG 212
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQTSHIA 120
++GNT IGLA+AS GYR V M +K G+ ++ P W +A
Sbjct 213 SAGNTAIGLALASKPRGYRVVCDM-----------LKVCGALVVEVPFHKVW-HPDHFVA 260
Query 121 LSLRL 125
S RL
Sbjct 261 FSGRL 265
> ath:AT3G61440 CYSC1; CYSC1 (CYSTEINE SYNTHASE C1); L-3-cyanoalanine
synthase/ cysteine synthase; K13034 L-3-cyanoalanine
synthase/ cysteine synthase [EC:2.5.1.47 4.4.1.9]
Length=272
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 40/70 (57%), Gaps = 1/70 (1%)
Query 41 MIRAAEASGHLAPG-CALIEPTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKAL 99
MI AE + PG LIEPTSGN GI LA +A+ GYR + MP S E+ M++
Sbjct 1 MIADAEKKKLIIPGKTTLIEPTSGNMGISLAFMAAMKGYRIIMTMPSYTSLERRVTMRSF 60
Query 100 GSTILRTPTA 109
G+ ++ T A
Sbjct 61 GAELVLTDPA 70
> cel:T25D3.3 hypothetical protein
Length=427
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 44/91 (48%), Gaps = 0/91 (0%)
Query 17 SCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPTSGNTGIGLAMASAVI 76
S +L+ K E S GS K R + ++ A GH+ G + E +SGNT L ++
Sbjct 77 SVRLVFKNESASLTGSLKHRYSWSLMMWALLEGHVKNGTHVFEASSGNTACSLGYMCRLL 136
Query 77 GYRSVAVMPMKMSTEKHRIMKALGSTILRTP 107
G + AV+P + K R ++ ++R P
Sbjct 137 GLQFTAVVPDTIEQVKVRRIEEQEGFVVRVP 167
> dre:100331432 serine racemase-like
Length=247
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 45/112 (40%), Gaps = 6/112 (5%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TPL+ + SC + K E + GS K RG+ A G G + +
Sbjct 6 TPLIPWCQTTLPPHTSCNIHIKMENMQRTGSFK---IRGV---ANQFGRRLKGDHFVTMS 59
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTAAAWD 113
SGN G A A G++ VM + ++K+LG + R PT D
Sbjct 60 SGNYGKAFAFACKHYGFKGKVVMLENAPISRSELIKSLGVEVERVPTTQLMD 111
> ath:AT3G10050 OMR1; OMR1 (L-O-METHYLTHREONINE RESISTANT 1);
L-threonine ammonia-lyase (EC:4.3.1.19); K01754 threonine dehydratase
[EC:4.3.1.19]
Length=592
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 55/103 (53%), Gaps = 5/103 (4%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
+PL +++ LG+ L K E L P S K R A M+ A LA G +I +
Sbjct 111 SPLQLAKKLSKRLGVRMYL--KREDLQPVFSFKLRGAYNMMVKLPAD-QLAKG--VICSS 165
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
+GN G+A++++ +G +V VMP+ K + ++ LG+T++
Sbjct 166 AGNHAQGVALSASKLGCTAVIVMPVTTPEIKWQAVENLGATVV 208
> cel:F01D4.8 hypothetical protein
Length=430
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 29/109 (26%), Positives = 49/109 (44%), Gaps = 4/109 (3%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHL-APGCALIEP 60
TPLV+ S + + K E +S + K R A ++ A G + + A+ +
Sbjct 59 TPLVKFSPPGFP---NTDIFFKNETVSATRTLKHRFAWALLLWAIVEGKVTSKTSAVYDA 115
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPTA 109
TSGN G A ++G AV+ + EK ++ G +++ PTA
Sbjct 116 TSGNMGSAEAYMCTLVGIPYFAVVSEECEQEKIDNIEKFGGKVIKVPTA 164
> dre:100331498 serine racemase-like
Length=323
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 41/107 (38%), Gaps = 6/107 (5%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPT 61
TP++ S+ L SC + K E + GS K R H + +
Sbjct 31 TPMIPWSQTTLPLHTSCNIHIKLENMQRTGSFKIRGVANQFAKRHKGDHF------VTMS 84
Query 62 SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTPT 108
+GN G + A G + VMP + +++ LG + R PT
Sbjct 85 AGNYGKAFSYACKHYGSKGKVVMPETAPISRSVLIQNLGVEVERVPT 131
> eco:b3772 ilvA, ECK3764, JW3745; threonine deaminase (EC:4.3.1.19);
K01754 threonine dehydratase [EC:4.3.1.19]
Length=514
Score = 33.1 bits (74), Expect = 0.21, Method: Composition-based stats.
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRA--AEASGHLAPGCALIE 59
TPL ++ ++++ L +L K E P S K R A M+ E H +I
Sbjct 32 TPLQKMEKLSSRL--DNVILVKREDRQPVHSFKLRGAYAMMAGLTEEQKAH-----GVIT 84
Query 60 PTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
++GN G+A +SA +G +++ VMP + K ++ G +L
Sbjct 85 ASAGNHAQGVAFSSARLGVKALIVMPTATADIKVDAVRGFGGEVL 129
> mmu:27364 Srr, Srs; serine racemase (EC:4.3.1.17 5.1.1.18 4.3.1.18);
K12235 serine racemase [EC:5.1.1.18]
Length=339
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 37/86 (43%), Gaps = 0/86 (0%)
Query 19 QLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPTSGNTGIGLAMASAVIGY 78
L KCE GS K R A IR A++ +SGN G L A+ + G
Sbjct 41 NLFFKCELFQKTGSFKIRGALNAIRGLIPDTPEEKPKAVVTHSSGNHGQALTYAAKLEGI 100
Query 79 RSVAVMPMKMSTEKHRIMKALGSTIL 104
+ V+P K ++A G++I+
Sbjct 101 PAYIVVPQTAPNCKKLAIQAYGASIV 126
> cel:T01H8.2 hypothetical protein
Length=317
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 46/104 (44%), Gaps = 6/104 (5%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARGMIRAA-EASGHLAPGCALIEP 60
T ++ + +G +L KCE L GS K ARG + +A A A G +I
Sbjct 24 TLVITSENIDEKVGNGTHVLFKCEHLQKTGSFK---ARGALNSAILAKEKNAKG--MIAH 78
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
+SGN G LA A+ IG V+P K M+ + I+
Sbjct 79 SSGNHGQALAWAAQKIGLPCTIVVPKNAPISKIEGMREYNANIV 122
> hsa:63826 SRR, ILV1, ISO1; serine racemase (EC:4.3.1.17 5.1.1.18
4.3.1.18); K12235 serine racemase [EC:5.1.1.18]
Length=340
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 38/86 (44%), Gaps = 0/86 (0%)
Query 19 QLLGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIEPTSGNTGIGLAMASAVIGY 78
L KCE GS K R A +R+ A++ +SGN G L A+ + G
Sbjct 41 NLFFKCELFQKTGSFKIRGALNAVRSLVPDALERKPKAVVTHSSGNHGQALTYAAKLEGI 100
Query 79 RSVAVMPMKMSTEKHRIMKALGSTIL 104
+ V+P K ++A G++I+
Sbjct 101 PAYIVVPQTAPDCKKLAIQAYGASIV 126
> cel:F13B12.4 hypothetical protein
Length=435
Score = 32.7 bits (73), Expect = 0.32, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 38/79 (48%), Gaps = 1/79 (1%)
Query 32 STKDRIARGMIRAAEASGHL-APGCALIEPTSGNTGIGLAMASAVIGYRSVAVMPMKMST 90
+ K R A ++ A G + + A+ + TSGNTG A ++ AV+ +
Sbjct 87 TLKHRFAWALLLWAITEGKVTSKTSAVYDSTSGNTGSAEAYMCTLVNVPYYAVVADNLEK 146
Query 91 EKHRIMKALGSTILRTPTA 109
EK + +++ G I++ P A
Sbjct 147 EKVKQIESFGGKIIKVPVA 165
> cel:K01C8.1 hypothetical protein; K01754 threonine dehydratase
[EC:4.3.1.19]
Length=499
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 50/104 (48%), Gaps = 7/104 (6%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIAR-GMIRAAEASGHLAPGCALIEP 60
TP VR ++++ + L K E+L GS K+R AR + + AE +I
Sbjct 118 TPCVRSLQLSSKCEMD--LYFKKEYLQVTGSFKERGARYALSKMAEQFKK----AGVIAA 171
Query 61 TSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
++GN + L+ +G VMP+ K + ++LG+T++
Sbjct 172 SAGNHALALSYHGQQMGIPVTVVMPVIAPLMKIQFCRSLGATVI 215
> mmu:69956 Ptcd3, 2610034F17Rik, 2810422B04Rik, AA589622, AU045708;
pentatricopeptide repeat domain 3
Length=685
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 7/65 (10%)
Query 54 GCALIEPT------SGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTILRTP 107
G + EP +G + S + G + V+P K + +K +++AL ST+ R P
Sbjct 25 GTGVCEPKVCCRFYAGTESLPKVEGSDITGIEEI-VIPKKKTWDKVAVLQALASTVNRDP 83
Query 108 TAAAW 112
TAA +
Sbjct 84 TAAPY 88
> ath:AT5G38530 tryptophan synthase-related; K06001 tryptophan
synthase beta chain [EC:4.2.1.20]
Length=506
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 50/112 (44%), Gaps = 10/112 (8%)
Query 2 TPLVRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIA--RGMIRAAEASGHLAPGCALIE 59
TPL+R R+ L ++ K E SP GS K A + A E ++ + E
Sbjct 134 TPLIRAKRLEKLLQTPARIYFKYEGGSPAGSHKPNTAVPQAYYNAKEGVKNV-----VTE 188
Query 60 PTSGNTGIGLAMASAVIGYR-SVAVMPMKMSTEKHR--IMKALGSTILRTPT 108
+G G LA AS++ G V + T+ +R +M+ G+ + +P+
Sbjct 189 TGAGQWGSSLAFASSLFGLDCEVWQVANSYHTKPYRRLMMQTWGAKVHPSPS 240
> dre:553325 ptcd3, MGC123014, fa08d08, wu:fa08d08, zgc:123014;
pentatricopeptide repeat domain 3
Length=667
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 83 VMPMKMSTEKHRIMKALGSTILRTPTAAAWDEQ 115
V+P K + K +++AL ST+ R PTA+ + Q
Sbjct 48 VIPKKKTWSKEAVLQALASTVKRDPTASDYRLQ 80
> sce:YER086W ILV1, ISO1; Ilv1p (EC:4.3.1.19); K01754 threonine
dehydratase [EC:4.3.1.19]
Length=576
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 28/105 (26%), Positives = 51/105 (48%), Gaps = 7/105 (6%)
Query 4 LVRLSRVAAALGISCQL----LGKCEFLSPGGSTKDRIARGMIRAAEASGHLAPGCALIE 59
++ S ++ +G+S +L + K E L P S K R A MI + S +I
Sbjct 75 VINESPISQGVGLSSRLNTNVILKREDLLPVFSFKLRGAYNMIAKLDDSQR---NQGVIA 131
Query 60 PTSGNTGIGLAMASAVIGYRSVAVMPMKMSTEKHRIMKALGSTIL 104
++GN G+A A+ + + VMP+ + K++ + LGS ++
Sbjct 132 CSAGNHAQGVAFAAKHLKIPATIVMPVCTPSIKYQNVSRLGSQVV 176
> ath:AT5G51230 EMF2; EMF2 (EMBRYONIC FLOWER 2); DNA binding /
transcription factor
Length=626
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Query 5 VRLSRVAAALGISCQLLGKCEFLSPGGSTKDRIARG----MIRAAEASGHLAPGCA 56
VR SR LG+ C++L K + S K RI G I AE+ + PG +
Sbjct 399 VRSSRQGPHLGLGCEVLDKTDDAHSVRSEKSRIPPGKHYERIGGAESGQRVPPGTS 454
Lambda K H
0.320 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40