bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1565_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
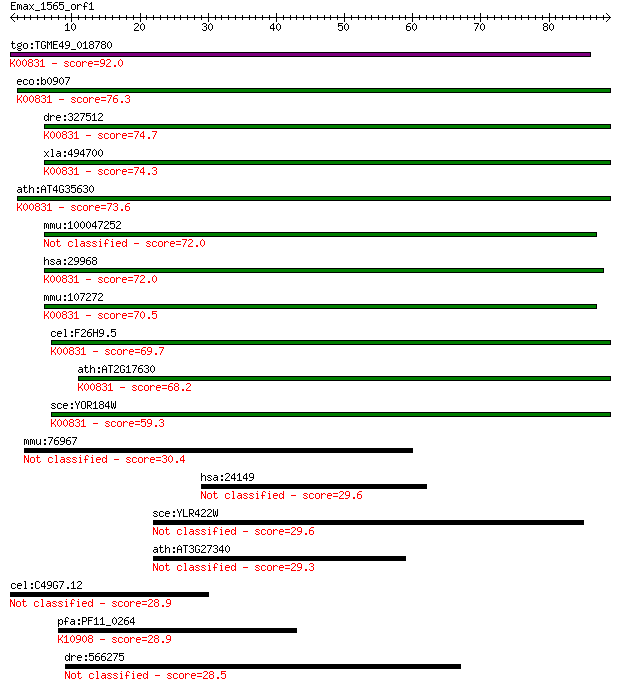
tgo:TGME49_018780 phosphoserine aminotransferase, putative (EC... 92.0 4e-19
eco:b0907 serC, ECK0898, JW0890, pdxC, pdxF; 3-phosphoserine/p... 76.3 2e-14
dre:327512 psat1, MGC77622, fi15b02, wu:fi15b02, zgc:55738, zg... 74.7 7e-14
xla:494700 psat1; phosphoserine aminotransferase 1 (EC:2.6.1.5... 74.3 9e-14
ath:AT4G35630 PSAT; PSAT; O-phospho-L-serine:2-oxoglutarate am... 73.6 2e-13
mmu:100047252 phosphoserine aminotransferase-like 72.0 4e-13
hsa:29968 PSAT1, EPIP, MGC1460, PSA, PSAT; phosphoserine amino... 72.0 4e-13
mmu:107272 Psat1, D8Ertd814e, EPIP, PSA, Psat; phosphoserine a... 70.5 1e-12
cel:F26H9.5 hypothetical protein; K00831 phosphoserine aminotr... 69.7 2e-12
ath:AT2G17630 phosphoserine aminotransferase, putative (EC:2.6... 68.2 6e-12
sce:YOR184W SER1, ADE9; 3-phosphoserine aminotransferase, cata... 59.3 3e-09
mmu:76967 2700049A03Rik, MGC183655, Talpid3, mKIAA0586; RIKEN ... 30.4 1.4
hsa:24149 ZNF318, FLJ21852, TZF, ZFP318; zinc finger protein 318 29.6
sce:YLR422W Protein of unknown function with similarity to hum... 29.6 2.6
ath:AT3G27340 hypothetical protein 29.3 3.0
cel:C49G7.12 hypothetical protein 28.9 3.9
pfa:PF11_0264 DNA-dependent RNA polymerase; K10908 DNA-directe... 28.9 4.2
dre:566275 bZ1P14.10, fj51b01, wu:fj51b01; si:rp71-1p14.10 28.5 4.8
> tgo:TGME49_018780 phosphoserine aminotransferase, putative (EC:2.6.1.52);
K00831 phosphoserine aminotransferase [EC:2.6.1.52]
Length=437
Score = 92.0 bits (227), Expect = 4e-19, Method: Composition-based stats.
Identities = 43/85 (50%), Positives = 58/85 (68%), Gaps = 4/85 (4%)
Query 1 DIQPFTDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQI 60
D+ P ++WTLN NAAY+HYCDNET++G+EFP P+ S L L +D +SN ++ I
Sbjct 149 DLPPVSEWTLNPNAAYVHYCDNETVYGVEFPRTPNLSEAQL----LLTSDMSSNFCSRPI 204
Query 61 PWNIIDLAYAGAQKNIGIAGVTLVV 85
+ L AGAQKNIG AGVT+V+
Sbjct 205 DIDAHALIVAGAQKNIGPAGVTIVI 229
> eco:b0907 serC, ECK0898, JW0890, pdxC, pdxF; 3-phosphoserine/phosphohydroxythreonine
aminotransferase (EC:2.6.1.52); K00831
phosphoserine aminotransferase [EC:2.6.1.52]
Length=362
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/87 (42%), Positives = 56/87 (64%), Gaps = 5/87 (5%)
Query 2 IQPFTDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIP 61
++P +W L+ NAAY+HYC NETI G+ PDF + + + ADF+S + ++ I
Sbjct 131 VKPMREWQLSDNAAYMHYCPNETIDGIAIDETPDFGADVV-----VAADFSSTILSRPID 185
Query 62 WNIIDLAYAGAQKNIGIAGVTLVVVHK 88
+ + YAGAQKNIG AG+T+V+V +
Sbjct 186 VSRYGVIYAGAQKNIGPAGLTIVIVRE 212
> dre:327512 psat1, MGC77622, fi15b02, wu:fi15b02, zgc:55738,
zgc:77622; phosphoserine aminotransferase 1 (EC:2.6.1.52); K00831
phosphoserine aminotransferase [EC:2.6.1.52]
Length=368
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 35/83 (42%), Positives = 54/83 (65%), Gaps = 6/83 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W+LN +A+Y++YC NET+HG+EF PD LV+D +SN ++ + +
Sbjct 137 STWSLNPSASYVYYCCNETVHGVEFNFIPDTKGV------ILVSDMSSNFLSRPVDVSKF 190
Query 66 DLAYAGAQKNIGIAGVTLVVVHK 88
L +AGAQKN+G AGVT+V+V +
Sbjct 191 GLIFAGAQKNVGCAGVTVVIVRE 213
> xla:494700 psat1; phosphoserine aminotransferase 1 (EC:2.6.1.52);
K00831 phosphoserine aminotransferase [EC:2.6.1.52]
Length=369
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 35/83 (42%), Positives = 50/83 (60%), Gaps = 6/83 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN A+Y++YC NET+HG+EF PD LV D +SN +K + +
Sbjct 138 SSWNLNPEASYVYYCANETVHGVEFQKVPDVKG------AVLVCDMSSNFLSKPVDVSKF 191
Query 66 DLAYAGAQKNIGIAGVTLVVVHK 88
L +AGAQKN+G AGVT+ +V +
Sbjct 192 GLIFAGAQKNVGCAGVTVAIVRE 214
> ath:AT4G35630 PSAT; PSAT; O-phospho-L-serine:2-oxoglutarate
aminotransferase (EC:2.6.1.52); K00831 phosphoserine aminotransferase
[EC:2.6.1.52]
Length=430
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 6/87 (6%)
Query 2 IQPFTDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIP 61
+ F + +A YLH C NETIHG+EF +P +P +LVAD +SN +K +
Sbjct 199 VPSFEELEQTPDAKYLHICANETIHGVEFKDYP------VPKNGFLVADMSSNFCSKPVD 252
Query 62 WNIIDLAYAGAQKNIGIAGVTLVVVHK 88
+ + Y GAQKN+G +GVT+V++ K
Sbjct 253 VSKFGVIYGGAQKNVGPSGVTIVIIRK 279
> mmu:100047252 phosphoserine aminotransferase-like
Length=370
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 33/81 (40%), Positives = 52/81 (64%), Gaps = 6/81 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN++A+Y+++C NET+HG+EF PD LV D +SN ++ + +
Sbjct 138 STWNLNRDASYVYFCANETVHGVEFDFVPDVKG------AVLVCDMSSNFLSRPVDVSKF 191
Query 66 DLAYAGAQKNIGIAGVTLVVV 86
+ +AGAQKN+G AGVT+V+V
Sbjct 192 GVIFAGAQKNVGSAGVTVVIV 212
> hsa:29968 PSAT1, EPIP, MGC1460, PSA, PSAT; phosphoserine aminotransferase
1 (EC:2.6.1.52); K00831 phosphoserine aminotransferase
[EC:2.6.1.52]
Length=324
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/82 (42%), Positives = 51/82 (62%), Gaps = 6/82 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN +A+Y++YC NET+HG+EF PD LV D +SN +K + +
Sbjct 138 STWNLNPDASYVYYCANETVHGVEFDFIPDVKG------AVLVCDMSSNFLSKPVDVSKF 191
Query 66 DLAYAGAQKNIGIAGVTLVVVH 87
+ +AGAQKN+G AGVT+V+V
Sbjct 192 GVIFAGAQKNVGSAGVTVVIVR 213
> mmu:107272 Psat1, D8Ertd814e, EPIP, PSA, Psat; phosphoserine
aminotransferase 1 (EC:2.6.1.52); K00831 phosphoserine aminotransferase
[EC:2.6.1.52]
Length=367
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 33/81 (40%), Positives = 51/81 (62%), Gaps = 6/81 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN +A+Y+++C NET+HG+EF PD LV D +SN ++ + +
Sbjct 135 STWNLNPDASYVYFCANETVHGVEFDFVPDVKG------AVLVCDMSSNFLSRPVDVSKF 188
Query 66 DLAYAGAQKNIGIAGVTLVVV 86
+ +AGAQKN+G AGVT+V+V
Sbjct 189 GVIFAGAQKNVGSAGVTVVIV 209
> cel:F26H9.5 hypothetical protein; K00831 phosphoserine aminotransferase
[EC:2.6.1.52]
Length=370
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 36/83 (43%), Positives = 55/83 (66%), Gaps = 7/83 (8%)
Query 7 DWTLNKNAAYLHYCDNETIHGLEF-PSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+W ++ AAYL+YC NET+HG+EF P+ P+ S ++P LVAD +SN + +
Sbjct 137 NWVHDEKAAYLYYCANETVHGIEFTPTAPE--SHNVP----LVADVSSNFMARPFDFKDH 190
Query 66 DLAYAGAQKNIGIAGVTLVVVHK 88
+ + GAQKN+G AG+T+V+V K
Sbjct 191 GVVFGGAQKNLGAAGLTIVIVRK 213
> ath:AT2G17630 phosphoserine aminotransferase, putative (EC:2.6.1.52);
K00831 phosphoserine aminotransferase [EC:2.6.1.52]
Length=422
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 34/79 (43%), Positives = 51/79 (64%), Gaps = 6/79 (7%)
Query 11 NKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPR-YLVADFASNLGTKQIPWNIIDLAY 69
+ +A YLH C NETIHG+EF +P + +P L+AD +SN +K + + + Y
Sbjct 198 SSDAKYLHICANETIHGVEFKDYP-----LVENPDGVLIADMSSNFCSKPVDVSKFGVIY 252
Query 70 AGAQKNIGIAGVTLVVVHK 88
AGAQKN+G +GVT+V++ K
Sbjct 253 AGAQKNVGPSGVTIVIIRK 271
> sce:YOR184W SER1, ADE9; 3-phosphoserine aminotransferase, catalyzes
the formation of phosphoserine from 3-phosphohydroxypyruvate,
required for serine and glycine biosynthesis; regulated
by the general control of amino acid biosynthesis mediated
by Gcn4p (EC:2.6.1.52); K00831 phosphoserine aminotransferase
[EC:2.6.1.52]
Length=395
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 52/83 (62%), Gaps = 4/83 (4%)
Query 7 DWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRY-LVADFASNLGTKQIPWNII 65
D K +Y++ C+NET+HG+E+P P + P +VAD +S++ +++I +
Sbjct 153 DKIKGKAFSYVYLCENETVHGVEWPELP---KCLVNDPNIEIVADLSSDILSRKIDVSQY 209
Query 66 DLAYAGAQKNIGIAGVTLVVVHK 88
+ AGAQKNIG+AG+TL ++ K
Sbjct 210 GVIMAGAQKNIGLAGLTLYIIKK 232
> mmu:76967 2700049A03Rik, MGC183655, Talpid3, mKIAA0586; RIKEN
cDNA 2700049A03 gene
Length=1520
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 8/57 (14%)
Query 3 QPFTDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQ 59
QP W L K + ET+H FPS + + +P P+Y D +LG K+
Sbjct 403 QPKMGWNLEKRDS------TETLHSQIFPSSEERGTAQVPVPKY--NDVVHDLGQKK 451
> hsa:24149 ZNF318, FLJ21852, TZF, ZFP318; zinc finger protein
318
Length=2279
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 1/33 (3%)
Query 29 EFPSFPDFSSFSLPSPRYLVADFASNLGTKQIP 61
+F P FS F L SP+ LV DF + G + P
Sbjct 2061 DFEPIPSFSGFPLDSPKTLVLDFETE-GERNSP 2092
> sce:YLR422W Protein of unknown function with similarity to human
DOCK proteins (guanine nucleotide exchange factors); interacts
with Ino4p; green fluorescent protein (GFP)-fusion protein
localizes to the cytoplasm, YLR422W is not an essential
protein
Length=1932
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 30/67 (44%), Gaps = 6/67 (8%)
Query 22 NETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII----DLAYAGAQKNIG 77
N+T+ + FP P+ SSF YL+ + A N Q P + DL A + N
Sbjct 1455 NDTLEAISFPPLPEQSSFE--RKEYLLKESARNFSRGQKPEKALAVYKDLIKAYDEINYD 1512
Query 78 IAGVTLV 84
+ G+ V
Sbjct 1513 LNGLAFV 1519
> ath:AT3G27340 hypothetical protein
Length=96
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 22 NETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTK 58
+ +HG F + P S FS+ SP+ + ++A GTK
Sbjct 9 SRRVHGTSFITIPKLSGFSIVSPKQVEVEYAD--GTK 43
> cel:C49G7.12 hypothetical protein
Length=287
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 1 DIQPFTDWTLNKNAAYLHYCDNETIHGLE 29
D+ PF ++LNK + CD ETI LE
Sbjct 59 DLTPFVKYSLNKKRRWETKCDLETISQLE 87
> pfa:PF11_0264 DNA-dependent RNA polymerase; K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1531
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 2/35 (5%)
Query 8 WTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLP 42
W +NK LHY + IHG+ P + +++LP
Sbjct 970 WKINKEI--LHYIEYAYIHGITIGKIPLYKNYTLP 1002
> dre:566275 bZ1P14.10, fj51b01, wu:fj51b01; si:rp71-1p14.10
Length=857
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Query 9 TLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPS--PRYLVADFASNLGTKQIPWNIID 66
T ++A L+ C E H L FP FS+ PS P +L A F G IP N ++
Sbjct 799 TTGRDAKALYSCQAEHSHELSFPQGAHFSNV-YPSVEPGWLQATFDGKTGL--IPENYVE 855
Lambda K H
0.321 0.138 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2021645584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40