bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1545_orf1
Length=126
Score E
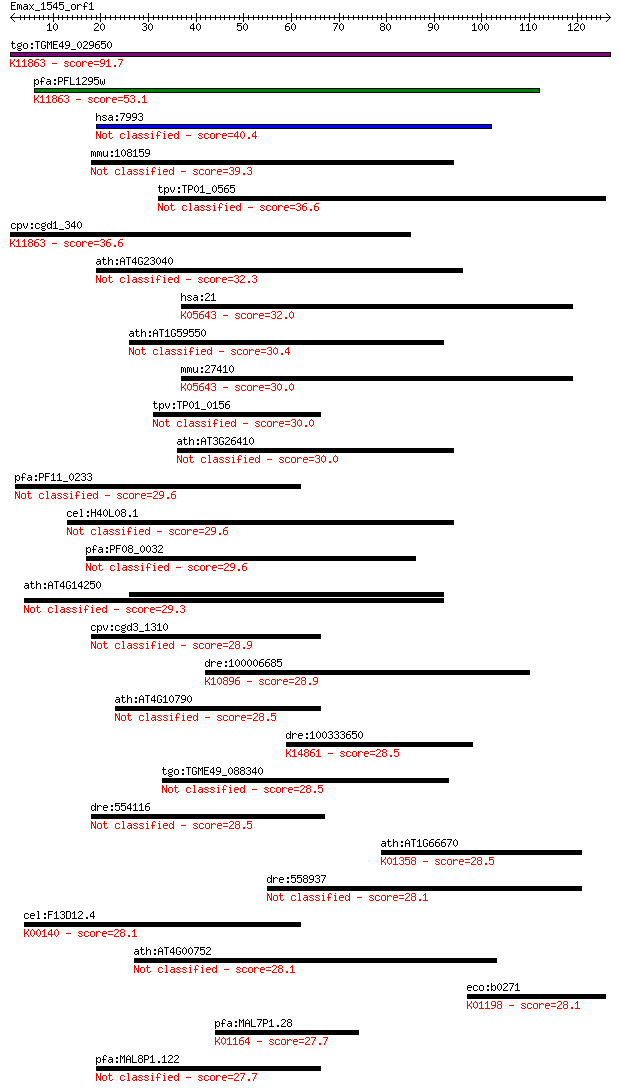
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_029650 machado-joseph disease protein, putative ; K... 91.7 5e-19
pfa:PFL1295w conserved Plasmodium protein; K11863 Ataxin-3 [EC... 53.1 2e-07
hsa:7993 UBXN8, D8S2298E, REP8, UBXD6; UBX domain protein 8 40.4
mmu:108159 Ubxn8, D0H8S2298E, Rep-8, Rep8h, Ubxd6; UBX domain ... 39.3 0.003
tpv:TP01_0565 hypothetical protein 36.6 0.020
cpv:cgd1_340 N-terminal machado-Joseph disease protein like do... 36.6 0.023
ath:AT4G23040 UBX domain-containing protein 32.3 0.36
hsa:21 ABCA3, ABC-C, ABC3, EST111653, LBM180, MGC166979, MGC72... 32.0 0.52
ath:AT1G59550 UBX domain-containing protein 30.4 1.5
mmu:27410 Abca3, 1810036E22Rik, ABC-C, Abc3, MGC90532; ATP-bin... 30.0 1.7
tpv:TP01_0156 hypothetical protein 30.0 1.7
ath:AT3G26410 methyltransferase/ nucleic acid binding 30.0 1.7
pfa:PF11_0233 conserved Plasmodium protein 29.6 2.3
cel:H40L08.1 hypothetical protein 29.6 2.4
pfa:PF08_0032 DNAJ protein, putative 29.6 2.5
ath:AT4G14250 structural constituent of ribosome 29.3 3.5
cpv:cgd3_1310 hypothetical protein 28.9 3.9
dre:100006685 fancm; Fanconi anemia, complementation group M; ... 28.9 4.0
ath:AT4G10790 UBX domain-containing protein 28.5 4.8
dre:100333650 URB1 ribosome biogenesis 1 homolog; K14861 nucle... 28.5 5.4
tgo:TGME49_088340 UBX domain-containing protein (EC:3.6.3.14) 28.5 5.7
dre:554116 im:7151693; zgc:112045 28.5 6.0
ath:AT1G66670 CLPP3; CLPP3; serine-type endopeptidase; K01358 ... 28.5 6.1
dre:558937 dhx32; DEAH (Asp-Glu-Ala-His) box polypeptide 32 28.1
cel:F13D12.4 alh-8; ALdehyde deHydrogenase family member (alh-... 28.1 6.6
ath:AT4G00752 UBX domain-containing protein 28.1 7.2
eco:b0271 yagH, ECK0272, JW0264; CP4-6 prophage; predicted xyl... 28.1 7.6
pfa:MAL7P1.28 ribonucleases p/mrp protein subunit, putative (E... 27.7 8.9
pfa:MAL8P1.122 Ubiquitin regulatory protein, putative 27.7 9.6
> tgo:TGME49_029650 machado-joseph disease protein, putative ;
K11863 Ataxin-3 [EC:3.4.22.-]
Length=412
Score = 91.7 bits (226), Expect = 5e-19, Method: Composition-based stats.
Identities = 49/126 (38%), Positives = 77/126 (61%), Gaps = 4/126 (3%)
Query 1 NEDPELQEAIRLSMECYRKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQ 60
++DPEL+EA+RLS+E Y++E++AP EEP + + I I +R G+++T++F +
Sbjct 281 DDDPELREALRLSLETYKQEMQAPPEEPPADAQNICTIVVRLKNGEKVTRRFRQTDTMEH 340
Query 61 IFKWVIYESHNNEGISPLTALSSFSLAQTLPRRRFCWTNGRIFLFVPPNTEGEDITEITL 120
+F+W YE+ +G T S L QT+P+R+FC G+I L +TEG I + L
Sbjct 341 VFQWAEYETSQRDGT---TLGRSCVLVQTVPKRKFCKLGGQICL-CEGDTEGIPIKDREL 396
Query 121 GGLGFQ 126
LGFQ
Sbjct 397 EELGFQ 402
> pfa:PFL1295w conserved Plasmodium protein; K11863 Ataxin-3 [EC:3.4.22.-]
Length=381
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 40/110 (36%), Positives = 59/110 (53%), Gaps = 9/110 (8%)
Query 6 LQEAIRLSMECYRKEVEAPGEEPTGEGEEIKLINL--RFPTGKRITKKFTKDVNTTQIFK 63
L+ A++LSME Y K + P EE + E IN+ + P K+I K+F+ + +F
Sbjct 256 LKIALKLSMEEYIKNMVPPLEETSDE----NCINVMVKLPN-KKIHKRFSFTKTLSDLFY 310
Query 64 WVIYESHNNEGI-SPLTALSSFSLAQTLPRRRFC-WTNGRIFLFVPPNTE 111
W+ YES + I S L +++ L Q PRR+FC + NG I L TE
Sbjct 311 WIEYESAQGQDITSSLLFKNNYYLYQLYPRRKFCKYQNGSIELHTGGKTE 360
> hsa:7993 UBXN8, D8S2298E, REP8, UBXD6; UBX domain protein 8
Length=270
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 41/83 (49%), Gaps = 9/83 (10%)
Query 19 KEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPL 78
KE+ EEP+ EE+ + LR P+G + ++F K ++ +F W+ ++
Sbjct 175 KEIPDLPEEPSQTAEEVVTVALRCPSGNVLRRRFLKSYSSQVLFDWMTRIGYH------- 227
Query 79 TALSSFSLAQTLPRRRFCWTNGR 101
+S +SL+ + PRR G+
Sbjct 228 --ISLYSLSTSFPRRPLAVEGGQ 248
> mmu:108159 Ubxn8, D0H8S2298E, Rep-8, Rep8h, Ubxd6; UBX domain
protein 8
Length=277
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 38/76 (50%), Gaps = 9/76 (11%)
Query 18 RKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISP 77
RKEV EEP+ EE+ + LR P G+ + ++F K N+ + W++ ++
Sbjct 180 RKEVPDLPEEPSETAEEVVTVALRCPNGRVLRRRFFKSWNSQVLLDWMMKVGYHK----- 234
Query 78 LTALSSFSLAQTLPRR 93
S + L+ + PRR
Sbjct 235 ----SLYRLSNSFPRR 246
> tpv:TP01_0565 hypothetical protein
Length=231
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 45/95 (47%), Gaps = 9/95 (9%)
Query 32 GEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSSFSLAQTLP 91
G + I ++ T +R+TK F+ + + ++F+W+ E L + L QT P
Sbjct 134 GSDAIRIAIKLITNERLTKSFSPNHSINKLFEWL-------ETKCDLKEHDFYFLIQTAP 186
Query 92 RRRFC-WTNGRIFLFVPPNTEGEDITEITLGGLGF 125
R+F +TNG I L N+ E +T T L F
Sbjct 187 YRKFIKYTNGSIELLTNSNSPRE-VTNETFNKLDF 220
> cpv:cgd1_340 N-terminal machado-Joseph disease protein like
domain, C-terminal UBX, DNA repair like domain ; K11863 Ataxin-3
[EC:3.4.22.-]
Length=397
Score = 36.6 bits (83), Expect = 0.023, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 41/87 (47%), Gaps = 4/87 (4%)
Query 1 NEDPELQEAIRLSMECYRKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQ 60
+E+ EL++ +R S + K + P E P I+ I +R G ++F K +
Sbjct 267 SEEKELEKVLRESAIEFAKSIPLPDEPPADHVNSIQ-IRVRSKVGSSFVRRFDKTNSCKH 325
Query 61 IFKWVIYESH---NNEGISPLTALSSF 84
+F W+ YE N+ SP + +S F
Sbjct 326 LFSWIEYEMALLGNSIHGSPYSFVSQF 352
> ath:AT4G23040 UBX domain-containing protein
Length=525
Score = 32.3 bits (72), Expect = 0.36, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 33/77 (42%), Gaps = 9/77 (11%)
Query 19 KEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPL 78
KE P E P GE I L +R P G R ++F K +F ++ I +
Sbjct 432 KEASLPQEPPAGEENAITL-QVRLPDGTRHGRRFFKSDKLQSLFDFI--------DICRV 482
Query 79 TALSSFSLAQTLPRRRF 95
+++ L + PRR F
Sbjct 483 VKPNTYRLVRPYPRRAF 499
> hsa:21 ABCA3, ABC-C, ABC3, EST111653, LBM180, MGC166979, MGC72201,
SMDP3; ATP-binding cassette, sub-family A (ABC1), member
3; K05643 ATP-binding cassette, subfamily A (ABC1), member
3
Length=1704
Score = 32.0 bits (71), Expect = 0.52, Method: Composition-based stats.
Identities = 24/86 (27%), Positives = 36/86 (41%), Gaps = 7/86 (8%)
Query 37 LINLR---FPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSSFSLAQTLPRR 93
+IN+R FP+ K D ++ + V++E N PL + L + RR
Sbjct 102 VINMRVRGFPSEKDFEDYIRYDNCSSSVLAAVVFEHPFNHSKEPLPLAVKYHLRFSYTRR 161
Query 94 RFCWTN-GRIFLFVPPNTEGEDITEI 118
+ WT G FL TEG T +
Sbjct 162 NYMWTQTGSFFL---KETEGWHTTSL 184
> ath:AT1G59550 UBX domain-containing protein
Length=307
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 9/68 (13%)
Query 26 EEPTGEGEE--IKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSS 83
+EP G+ + + I++RFP G+R +KF K Q+ W SH +E + +
Sbjct 218 KEPKGDCDRSVVCSISVRFPNGRRKQRKFLKS-EPVQLL-WSFCYSHMDESDNK-----A 270
Query 84 FSLAQTLP 91
F L Q +P
Sbjct 271 FKLVQAIP 278
> mmu:27410 Abca3, 1810036E22Rik, ABC-C, Abc3, MGC90532; ATP-binding
cassette, sub-family A (ABC1), member 3; K05643 ATP-binding
cassette, subfamily A (ABC1), member 3
Length=1704
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 36/86 (41%), Gaps = 7/86 (8%)
Query 37 LINLR---FPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSSFSLAQTLPRR 93
+I +R F + K D +++ + V++E N PL + L + RR
Sbjct 102 MIKMRVHGFSSEKDFEDYIRYDNHSSSVLAAVVFEHSFNHSQDPLPLAVKYHLRFSYTRR 161
Query 94 RFCWTN-GRIFLFVPPNTEGEDITEI 118
+ WT G IFL TEG T +
Sbjct 162 NYMWTQTGNIFL---KETEGWHTTSL 184
> tpv:TP01_0156 hypothetical protein
Length=340
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 31 EGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWV 65
E +E+K I +R P G RI +F K+ +I++WV
Sbjct 249 ERKEVK-IRVRLPNGNRIEGEFAKNDKVEKIYEWV 282
> ath:AT3G26410 methyltransferase/ nucleic acid binding
Length=477
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 13/58 (22%), Positives = 32/58 (55%), Gaps = 0/58 (0%)
Query 36 KLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSSFSLAQTLPRR 93
+L++ R P + + + F +++ + +W + E+H+N+ LSS +A+ + +R
Sbjct 10 RLLDFRKPEVEALAELFGEEIAENESLQWRLPENHHNDTPFHFVQLSSEEIARNIAKR 67
> pfa:PF11_0233 conserved Plasmodium protein
Length=1956
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 2 EDPELQEAIRLSMECYRKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQI 61
E+ + IR S + + +E E P EPT I I++ PT + + +K T ++ +
Sbjct 1393 EETTDKNMIRYSFDSFNEEFENPISEPTPAASIISYISVMSPTNEVLKEKNTSNMKNEHM 1452
> cel:H40L08.1 hypothetical protein
Length=444
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 41/81 (50%), Gaps = 14/81 (17%)
Query 13 SMECYRKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNN 72
++E Y++ + TG + + +RFPTGK++ KF D N +IF E+ +
Sbjct 331 TVETYKRTISGDTTLSTGSHDLL----IRFPTGKKVI-KFNADDNIEKIFD----EAMKS 381
Query 73 EGISPLTALSSFSLAQTLPRR 93
E + PL F + Q+ P++
Sbjct 382 E-MCPLF----FQMHQSFPKK 397
> pfa:PF08_0032 DNAJ protein, putative
Length=655
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 34/73 (46%), Gaps = 7/73 (9%)
Query 17 YRKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKW----VIYESHNN 72
Y K V G E T EE I + K +TKKF K +N I K+ + Y S+
Sbjct 480 YNKFVSKNGSEET---EEQLFIQPVYVLKKNLTKKFNKFINDKTINKYDSFLLDYSSNTF 536
Query 73 EGISPLTALSSFS 85
I+ + LS++S
Sbjct 537 SSINEIKNLSNYS 549
> ath:AT4G14250 structural constituent of ribosome
Length=724
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 9/68 (13%)
Query 26 EEPTGEGEEIKLINL--RFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSS 83
EEP G+ + + +L RFP G+R +KF K Q+ W SH +E + +
Sbjct 635 EEPKGDCDRSVVCSLCVRFPDGRRKQRKFLKS-EPIQLL-WSFCYSHIDE-----SEKKA 687
Query 84 FSLAQTLP 91
F L Q +P
Sbjct 688 FKLVQAIP 695
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 40/97 (41%), Gaps = 16/97 (16%)
Query 4 PELQEAIRLS-------MECYRKEVEAPGEEPTGEGEE--IKLINLRFPTGKRITKKFTK 54
PE ++ + LS + C E EEP + + + I +RFP G+R +KF K
Sbjct 263 PEFEDIMTLSEHEEETCLSCDLLEFPVLTEEPKADCDRSVVCSICVRFPDGRRKQRKFLK 322
Query 55 DVNTTQIFKWVIYESHNNEGISPLTALSSFSLAQTLP 91
Q+ W SH E + F L Q +P
Sbjct 323 S-EPIQLL-WSFCYSHMEE-----SEKKEFKLVQAIP 352
> cpv:cgd3_1310 hypothetical protein
Length=306
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 18 RKEVEAPGEEPTG-EGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWV 65
R++++A ++ E E + + ++ GK+ + F KD + +IFKW+
Sbjct 198 REKLQAEFQQDRDIEQESLSRVCVKNSAGKKFQRNFHKDDSVYEIFKWI 246
> dre:100006685 fancm; Fanconi anemia, complementation group M;
K10896 fanconi anemia group M protein [EC:3.6.4.13]
Length=1761
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 30/68 (44%), Gaps = 6/68 (8%)
Query 42 FPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSSFSLAQTLPRRRFCWTNGR 101
FP GK + TK + QI + GI P + + + P+RR W++ R
Sbjct 118 FPAGKIVFMAPTKPLVAQQI-----EACYRVMGI-PQEHTAELTGSTAAPQRRSLWSSRR 171
Query 102 IFLFVPPN 109
+F PP+
Sbjct 172 VFFLTPPD 179
> ath:AT4G10790 UBX domain-containing protein
Length=480
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Query 23 APGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWV 65
A GEEP +G ++ + +RFP G+R + F + ++ +V
Sbjct 392 ALGEEPE-KGPDVTQVLVRFPNGERKGRMFKSETKIQTLYDYV 433
> dre:100333650 URB1 ribosome biogenesis 1 homolog; K14861 nucleolar
pre-ribosomal-associated protein 1
Length=2273
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 59 TQIFKWVIYESHNNEGISPLTALSSFSLAQTLPRRRFCW 97
T+IF ++ ES + + P+ LA LP +F W
Sbjct 600 TEIFAGIVTESGMTQEVPPVLQYQILQLALELPASKFSW 638
> tgo:TGME49_088340 UBX domain-containing protein (EC:3.6.3.14)
Length=492
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 3/63 (4%)
Query 33 EEIKLINLRFPTGKRITKKFTKDVNTTQIFKWV--IYESHNNEGIS-PLTALSSFSLAQT 89
E+ I LR P+G R +++F + +++ W + E NE I PL +T
Sbjct 404 EQQTAICLRLPSGSRFSRQFPAQTSLEELYLWADCLAEFTENEEIDIPLNFHLVVPPRRT 463
Query 90 LPR 92
LPR
Sbjct 464 LPR 466
> dre:554116 im:7151693; zgc:112045
Length=272
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 18 RKEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVI 66
+K V +EP+ + E I LRFP + I ++F K ++ + +W++
Sbjct 175 QKRVIVLPDEPSVDTEGAVKIALRFPGRRAIHRRFLKTWSSQLLLEWMM 223
> ath:AT1G66670 CLPP3; CLPP3; serine-type endopeptidase; K01358
ATP-dependent Clp protease, protease subunit [EC:3.4.21.92]
Length=309
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 11/42 (26%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 79 TALSSFSLAQTLPRRRFCWTNGRIFLFVPPNTEGEDITEITL 120
++ +F LA +R+C N ++ + P T G TE+++
Sbjct 163 ASMGAFLLASGSKGKRYCMPNSKVMIHQPLGTAGGKATEMSI 204
> dre:558937 dhx32; DEAH (Asp-Glu-Ala-His) box polypeptide 32
Length=731
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 16/69 (23%), Positives = 32/69 (46%), Gaps = 12/69 (17%)
Query 55 DVNTTQIFKWVIYESHN---NEGISPLTALSSFSLAQTLPRRRFCWTNGRIFLFVPPNTE 111
+VN + +WV+Y H N I ++ +S+ Q P + F + P++E
Sbjct 639 EVNKKNLPEWVLYHEHTLSENNCIRTVSQISAIEFIQMAP---------QYFFYNLPSSE 689
Query 112 GEDITEITL 120
+D+ + T+
Sbjct 690 SKDLLQHTI 698
> cel:F13D12.4 alh-8; ALdehyde deHydrogenase family member (alh-8);
K00140 methylmalonate-semialdehyde dehydrogenase [EC:1.2.1.27]
Length=523
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 31/69 (44%), Gaps = 11/69 (15%)
Query 4 PELQEAIRLSMECYRKEVEAP---GEEPTGEGEEIKLINLR--------FPTGKRITKKF 52
P + ++ +M CYR+E+ P E E I++IN F + +KF
Sbjct 391 PTILAGVKPNMTCYREEIFGPVLVVMEAENLNEAIEIINNNPYGNGTAIFTSNGATARKF 450
Query 53 TKDVNTTQI 61
T +V+ QI
Sbjct 451 TNEVDVGQI 459
> ath:AT4G00752 UBX domain-containing protein
Length=469
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 17/76 (22%), Positives = 33/76 (43%), Gaps = 8/76 (10%)
Query 27 EPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWVIYESHNNEGISPLTALSSFSL 86
EP+GE E+ + +R P R ++F K +F ++ + L ++ +
Sbjct 382 EPSGENEDAITLLVRMPDSSRHGRRFLKSDKLKYLFDFI--------DAAGLVKPGTYRV 433
Query 87 AQTLPRRRFCWTNGRI 102
+ PRR F +G +
Sbjct 434 VRPYPRRAFSIQDGAL 449
> eco:b0271 yagH, ECK0272, JW0264; CP4-6 prophage; predicted xylosidase/arabinosidase;
K01198 xylan 1,4-beta-xylosidase [EC:3.2.1.37]
Length=536
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 15/29 (51%), Gaps = 0/29 (0%)
Query 97 WTNGRIFLFVPPNTEGEDITEITLGGLGF 125
W NGR FL P+ EG I +G GF
Sbjct 98 WKNGRNFLVTAPSIEGPWSEPIPMGNGGF 126
> pfa:MAL7P1.28 ribonucleases p/mrp protein subunit, putative
(EC:3.1.26.5); K01164 ribonuclease P/MRP protein subunit POP1
[EC:3.1.26.5]
Length=1156
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 44 TGKRITKKFTKDVNTTQIFKWVIYESHNNE 73
T K++ K F K +T ++ +WVI++ NN+
Sbjct 979 TLKQVIKFFIKHADTKKLLEWVIHKRINNK 1008
> pfa:MAL8P1.122 Ubiquitin regulatory protein, putative
Length=238
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 6/47 (12%)
Query 19 KEVEAPGEEPTGEGEEIKLINLRFPTGKRITKKFTKDVNTTQIFKWV 65
KE++ ++P I +++R GK+IT+KF D +F++V
Sbjct 155 KEIKIDDKKP------ITTLHIRLYNGKKITQKFNYDHTVEDLFQFV 195
Lambda K H
0.316 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40