bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1538_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
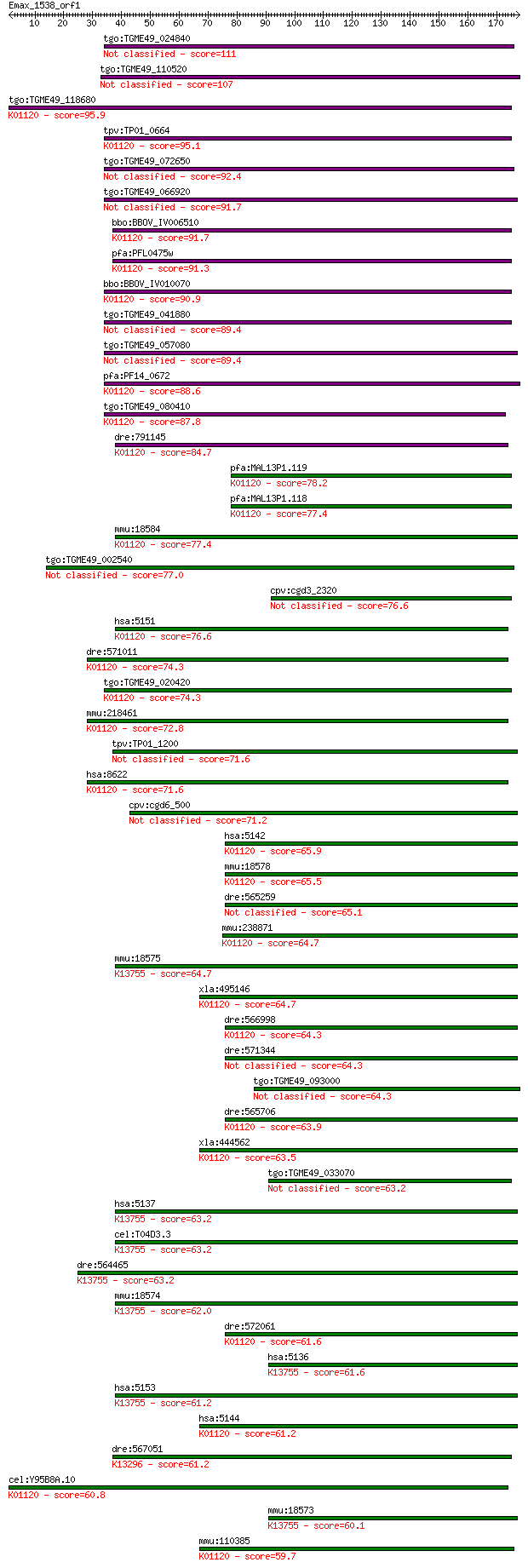
tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.1... 111 1e-24
tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic n... 107 3e-23
tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase, ... 95.9 7e-20
tpv:TP01_0664 3',5' cyclic nucleotide phosphodiesterase; K0112... 95.1 1e-19
tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic nu... 92.4 8e-19
tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17) 91.7 1e-18
bbo:BBOV_IV006510 23.m05980; 3'5'-cyclic nucleotide phosphodie... 91.7 1e-18
pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.1... 91.3 1e-18
bbo:BBOV_IV010070 23.m06154; 3'5'-cyclic nucleotide phosphodie... 90.9 2e-18
tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase, ... 89.4 6e-18
tgo:TGME49_057080 cAMP-specific 3',5'-cyclic phosphodiesterase... 89.4 6e-18
pfa:PF14_0672 cyclic nucleotide phosphodiesterase, putative; K... 88.6 1e-17
tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC... 87.8 2e-17
dre:791145 pde8a, zgc:158458; phosphodiesterase 8A (EC:3.1.4.1... 84.7 1e-16
pfa:MAL13P1.119 calcium/calmodulin-dependent 3',5'-cyclic nucl... 78.2 2e-14
pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:... 77.4 2e-14
mmu:18584 Pde8a, AI551852, Pde8; phosphodiesterase 8A (EC:3.1.... 77.4 2e-14
tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase, p... 77.0 3e-14
cpv:cgd3_2320 cGMP phosphodiesterase A4 76.6 4e-14
hsa:5151 PDE8A, FLJ16150, HsT19550; phosphodiesterase 8A (EC:3... 76.6 4e-14
dre:571011 IBMX-insensitive phosphodiesterase 8B-like; K01120 ... 74.3 2e-13
tgo:TGME49_020420 3'5'-cyclic nucleotide phosphodiesterase, pu... 74.3 2e-13
mmu:218461 Pde8b, B230331L10Rik, C030047E14Rik; phosphodiester... 72.8 6e-13
tpv:TP01_1200 hypothetical protein 71.6 1e-12
hsa:8622 PDE8B, ADSD, FLJ11212, PPNAD3; phosphodiesterase 8B (... 71.6 1e-12
cpv:cgd6_500 membrane associated HD superfamily cyclic nucleot... 71.2 2e-12
hsa:5142 PDE4B, DKFZp686F2182, DPDE4, MGC126529, PDE4B5, PDEIV... 65.9 7e-11
mmu:18578 Pde4b, Dpde4, R74983, dunce; phosphodiesterase 4B, c... 65.5 1e-10
dre:565259 pde4c, im:7160317, si:dkey-149i17.5; phosphodiester... 65.1 1e-10
mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D, ... 64.7 1e-10
mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755 ca... 64.7 2e-10
xla:495146 hypothetical LOC495146; K01120 3',5'-cyclic-nucleot... 64.7 2e-10
dre:566998 novel protein similar to vertebrate phosphodiestera... 64.3 2e-10
dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-25... 64.3 2e-10
tgo:TGME49_093000 3', 5'-cyclic nucleotide phosphodiesterase d... 64.3 2e-10
dre:565706 cAMP-specific 3,5-cyclic phosphodiesterase 4B-like;... 63.9 2e-10
xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specifi... 63.5 3e-10
tgo:TGME49_033070 phosphodiesterase, putative (EC:3.1.4.17) 63.2 4e-10
hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-depend... 63.2 4e-10
cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1); K1... 63.2 4e-10
dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent; ... 63.2 5e-10
mmu:18574 Pde1b, 63kDa, Pde1b1; phosphodiesterase 1B, Ca2+-cal... 62.0 1e-09
dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120 ... 61.6 1e-09
hsa:5136 PDE1A, HCAM1, HSPDE1A, MGC26303; phosphodiesterase 1A... 61.6 1e-09
hsa:5153 PDE1B, PDE1B1, PDES1B; phosphodiesterase 1B, calmodul... 61.2 2e-09
hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4... 61.2 2e-09
dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic ... 61.2 2e-09
cel:Y95B8A.10 pde-6; PhosphoDiEsterase family member (pde-6); ... 60.8 2e-09
mmu:18573 Pde1a, AI987702, AW125737, MGC116577; phosphodiester... 60.1 4e-09
mmu:110385 Pde4c, Dpde1, E130301F19Rik, MGC31320, dunce; phosp... 59.7 5e-09
> tgo:TGME49_024840 cAMP phosphodiesterase, putative (EC:3.1.4.17
1.1.99.3)
Length=1324
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 56/142 (39%), Positives = 79/142 (55%), Gaps = 1/142 (0%)
Query 34 IGLDWDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYH 93
+G+DWD D+L L+ + ++ VG+ LL V LGC +I+ L I+ +Y D PYH
Sbjct 686 VGVDWDFDMLHLNSQTENVIVEVGYALLCRLVPDLGCEEVRLIRFLHAIQMQYR-DNPYH 744
Query 94 TATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLA 153
H+A+VAH L L+ L +AAL HDVGH G+ N F + PLA
Sbjct 745 NKIHSAEVAHLTECLTRMLNAQRNMNSIDKVTLTVAALCHDVGHPGRNNQFFINAFDPLA 804
Query 154 TIYNDRSILENFHASVLFRIIN 175
IYND ++LENFH+ + FR +
Sbjct 805 VIYNDVAVLENFHSCLTFRTLE 826
> tgo:TGME49_110520 calcium/calmodulin-dependent 3', 5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.4.21.69 3.1.4.17)
Length=1531
Score = 107 bits (266), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 69/146 (47%), Positives = 90/146 (61%), Gaps = 4/146 (2%)
Query 33 GIGLDWDADILSLSQNYQGMLQRVGWVLLRPFV-SLLGCGNTPVIQLLQHIEARYDPDIP 91
IG WD D+LS + L VG VLL P + SL C + VI LLQ+++ARY + P
Sbjct 1159 AIGQVWDFDMLSFAALSPNPLVEVGLVLLLPEIDSLHCCTDQEVIVLLQNLQARYLQN-P 1217
Query 92 YHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHP 151
YH+ HAA+V H A L L P + + C +AA AHDVGH +TN FLQ HP
Sbjct 1218 YHSQVHAAEVVHTAACLMRCLI--PQRSAFANLCTLVAAAAHDVGHPARTNLFLQNLLHP 1275
Query 152 LATIYNDRSILENFHASVLFRIINEI 177
L+ +YND S LENFH+++LFRI++EI
Sbjct 1276 LSIVYNDVSTLENFHSALLFRILSEI 1301
> tgo:TGME49_118680 3',5'--cyclic-nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=812
Score = 95.9 bits (237), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 58/187 (31%), Positives = 88/187 (47%), Gaps = 14/187 (7%)
Query 1 RGKNRSAVHGKGSYESLGTAASQSFTSPPA------------FGGIGLDWDADILSLSQN 48
R A HG S++ A S P +G+D +IL S+
Sbjct 270 RSMVEKAEHGSSSFDDRVAAPKVSLELPKVTSLLATKYAVDLLPVVGIDISYNILEFSKK 329
Query 49 Y-QGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYHTATHAAQVAHAAMV 107
+LQ VG+VLL V GC + + + L ++ Y + YH H A VAH +
Sbjct 330 CPDTVLQEVGYVLLNRIVCDWGCEDKVLCEFLYMVKTLYREN-SYHNQIHGAMVAHYMVC 388
Query 108 LNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHA 167
L L ++ + +AAL HD+GH G+ N F S PLA +YND+++LENFH+
Sbjct 389 LLRGLGINREMNSLSTAACAVAALCHDIGHPGRNNNFFVASGAPLAVLYNDKAVLENFHS 448
Query 168 SVLFRII 174
++ FR++
Sbjct 449 ALTFRVL 455
> tpv:TP01_0664 3',5' cyclic nucleotide phosphodiesterase; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=450
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 63/151 (41%), Positives = 91/151 (60%), Gaps = 16/151 (10%)
Query 34 IGLDWDADILSLSQNYQGMLQR-----VGWVLLRPFVSLLGCG-NTPVIQLLQHIEARYD 87
IGL+W D++ LS NY +L+ VG LL +L+ NT +I +LQ+++ Y
Sbjct 107 IGLNWHLDLIELS-NYPTVLETGPIVVVGKHLLHCVGNLIHPNFNTNLIPVLQNLQQHYL 165
Query 88 PDIPYHTATHAAQVAHAAMVLN----TKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNG 143
+ PYH A HAA V H + +L+ TK L+P +E F +++LAHDVGH G+TN
Sbjct 166 AN-PYHNALHAATVGHMSKLLSNIVTTKRRLNPYEE----FAFIISSLAHDVGHPGKTNN 220
Query 144 FLQQSRHPLATIYNDRSILENFHASVLFRII 174
+L + + L+ IYND S LEN+H S+LF II
Sbjct 221 YLSNTSNVLSLIYNDNSTLENYHCSLLFYII 251
> tgo:TGME49_072650 calcium/calmodulin-dependent 3',5'-cyclic
nucleotide phosphodiesterase, putative (EC:3.1.4.17)
Length=1320
Score = 92.4 bits (228), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 58/157 (36%), Positives = 79/157 (50%), Gaps = 16/157 (10%)
Query 34 IGLDWDADILSLSQNYQG-MLQRVGWVLLRPFVS--LLGCGNTPVIQLLQHIEARYDPDI 90
+G+ W D+ L + G L VG+ LL P ++ L C V+ L ++ Y D
Sbjct 990 VGILWSLDLFKLDKECNGNALLHVGYQLLAPLLAAGYLTCSREVVLDFLYSLQCLY-IDT 1048
Query 91 PYHTATHAAQVAHAAMVLNTKLDL------DPGKEKTGS------FCLGLAALAHDVGHL 138
PYH HAA VAH A + LDL PG L +AAL HD GH
Sbjct 1049 PYHNQLHAASVAHLAFSICHFLDLFAPLENAPGDAAASQPVFAQYLSLAIAALGHDAGHP 1108
Query 139 GQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIIN 175
G++N FL Q+ L+ +YNDRSILEN+HA + F ++
Sbjct 1109 GRSNAFLVQTSSTLSVVYNDRSILENYHACLTFYTLS 1145
> tgo:TGME49_066920 phosphodiesterase, putative (EC:3.1.4.17)
Length=1065
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 52/143 (36%), Positives = 74/143 (51%), Gaps = 1/143 (0%)
Query 34 IGLDWDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYH 93
+G+ W+ D+L + + L G+VLL V+ GC + + L EA++ + PYH
Sbjct 556 VGVAWNFDLLEVEEETGHALSLTGFVLLNGVVADWGCPPSRLTNFLLSCEAQHQAN-PYH 614
Query 94 TATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLA 153
HAA VAHA L + + L +AAL HD+GH G+TN F S PLA
Sbjct 615 NQRHAAMVAHATAWLANTVGALNKCDSVERATLYVAALCHDLGHPGRTNQFFVSSHDPLA 674
Query 154 TIYNDRSILENFHASVLFRIINE 176
+YND S LEN H + FR + +
Sbjct 675 IVYNDISCLENLHCCLCFRTLQK 697
> bbo:BBOV_IV006510 23.m05980; 3'5'-cyclic nucleotide phosphodiesterase
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=777
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/138 (34%), Positives = 73/138 (52%), Gaps = 1/138 (0%)
Query 37 DWDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYHTAT 96
DW+ D+L + +G+ LL+ + G + V+ L IE +Y ++PYH
Sbjct 443 DWNFDVLEYFKKTPAGFMSIGYTLLQKYQEDYGIDRSVVVNFLYRIENQYR-NVPYHNKM 501
Query 97 HAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLATIY 156
H A VA + L + L L +AAL+HDVGH G+ N F ++ HP+A +Y
Sbjct 502 HGAMVAQKVLCLASYTGLLEHLSILDEALLMVAALSHDVGHPGRNNAFFVRTHHPVAQLY 561
Query 157 NDRSILENFHASVLFRII 174
ND S+LEN+HA+ R++
Sbjct 562 NDTSVLENYHAASALRVL 579
> pfa:PFL0475w PDE1; cGMP-specific phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=954
Score = 91.3 bits (225), Expect = 1e-18, Method: Composition-based stats.
Identities = 50/138 (36%), Positives = 74/138 (53%), Gaps = 2/138 (1%)
Query 37 DWDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYHTAT 96
DW+ +I ++ + +G+ LL P L + ++ Y DIPYHT+
Sbjct 626 DWNGNIENIYK--ANTFISIGYKLLYPLGVLEANFDKEKLKKFLFRICSYYNDIPYHTSL 683
Query 97 HAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLATIY 156
HAAQVAH + + LD++ FCL +++L HD GH G N FL S + LA Y
Sbjct 684 HAAQVAHFSKSMLFMLDMNHKISAIDEFCLHISSLCHDTGHPGLNNYFLINSENNLALTY 743
Query 157 NDRSILENFHASVLFRII 174
ND S+LEN+H S+LF+ +
Sbjct 744 NDNSVLENYHCSLLFKTL 761
> bbo:BBOV_IV010070 23.m06154; 3'5'-cyclic nucleotide phosphodiesterase
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=485
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 61/154 (39%), Positives = 87/154 (56%), Gaps = 22/154 (14%)
Query 34 IGLDWDADILSLSQNYQGMLQR-----VGWVLLRP--------FVSLLGCGNTPVIQLLQ 80
+G +W+ D ++S N++ +L+ VG L+ P F L+ TPV+ +Q
Sbjct 148 LGKNWNMDFFAMS-NFKFVLRSGPVVAVGHQLIDPVGDKIHPDFNKLI----TPVLNSIQ 202
Query 81 HIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQ 140
+ Y P+ PYH A H A VAH + VL L L+ F +AAL HD GH G+
Sbjct 203 DV---YLPN-PYHNALHGACVAHMSCVLLKALSLEQYLTPLEQFAYLIAALGHDAGHPGK 258
Query 141 TNGFLQQSRHPLATIYNDRSILENFHASVLFRII 174
TN FL+ +++PLA IYND SILEN+HAS++ II
Sbjct 259 TNAFLRSTQNPLALIYNDASILENYHASLVCHII 292
> tgo:TGME49_041880 3', 5'-cyclic nucleotide phosphodiesterase,
putative (EC:2.1.1.43 3.1.4.17)
Length=1281
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 75/142 (52%), Gaps = 2/142 (1%)
Query 34 IGLDWDADILSLSQNYQG-MLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPY 92
+ ++W+ ++ + Q +G L VG L+ V C + V ++ + Y P I Y
Sbjct 805 VSIEWNLSMIDVDQQCKGWCLYCVGSDLIFCKVQGFECEKSFVQNFMKVAQMCYQPTI-Y 863
Query 93 HTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPL 152
H H AQVAH + L KL+L L +AAL HD+GH G+ N F SR PL
Sbjct 864 HNHLHGAQVAHNTVWLARKLELASSLTAPELVALIVAALCHDIGHQGKNNAFYVASRSPL 923
Query 153 ATIYNDRSILENFHASVLFRII 174
IYND ++LENFHA + F+I+
Sbjct 924 GIIYNDIAVLENFHACLTFKIL 945
> tgo:TGME49_057080 cAMP-specific 3',5'-cyclic phosphodiesterase,
putative (EC:3.1.4.17)
Length=1691
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 56/149 (37%), Positives = 77/149 (51%), Gaps = 6/149 (4%)
Query 34 IGLDWDADILSLSQNYQG-MLQRVGWVLLRPFVSLLG--CGNTPVIQLLQHIEARYDPDI 90
IG+ WD D L++ L G V L P + G C V L+ ++ +Y P
Sbjct 1284 IGVAWDLDFFELNERVNNNALLVTGQVQLLPLLRPEGLRCSPQVVRCYLRCLQQQYCPSN 1343
Query 91 PYHTATHAAQVAHAAMVL-NTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSR 149
PYH HAA V+H +++ N L CL +A++AHDVGH G N +L S+
Sbjct 1344 PYHNQLHAAMVSHCCLIIVNEVLPSKQALTYVDELCLIIASVAHDVGHPGLNNQYLISSQ 1403
Query 150 HPLATIYNDRSILENFHASVLFRI--INE 176
LAT YND ++LEN+HA+ FR INE
Sbjct 1404 SLLATTYNDIAVLENYHAACCFRTAGINE 1432
> pfa:PF14_0672 cyclic nucleotide phosphodiesterase, putative;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=815
Score = 88.6 bits (218), Expect = 1e-17, Method: Composition-based stats.
Identities = 51/145 (35%), Positives = 74/145 (51%), Gaps = 2/145 (1%)
Query 34 IGLDWDADILSLSQNYQGM-LQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPY 92
IG +WD + + + VG+ L+ P++ + L I + Y P+ PY
Sbjct 400 IGKNWDYSFIDSEYGKSTLVILEVGYHLISPYIENNENKKKKLQLFLLLINSMYFPN-PY 458
Query 93 HTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPL 152
H A H A V H + L D D T C +A++AHDVGH G+TN +L ++ H L
Sbjct 459 HNANHGATVCHLSKCLAHITDYDSYLNNTYMICYLIASIAHDVGHPGKTNSYLSETNHIL 518
Query 153 ATIYNDRSILENFHASVLFRIINEI 177
+ YND SILEN+H S+ F I+ I
Sbjct 519 SIRYNDMSILENYHCSITFSILQLI 543
> tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC:3.1.4.17
1.6.5.3); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1085
Score = 87.8 bits (216), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 52/141 (36%), Positives = 74/141 (52%), Gaps = 3/141 (2%)
Query 34 IGLDWDADILSLSQNYQGMLQRVGWVLLRPF-VSLLGCGNTPVI-QLLQHIEARYDPDIP 91
IG WD ++ L L VG+VLL PF +S C NT ++ + L ++ Y + P
Sbjct 753 IGNSWDVNMFMLRLKLPRPLVEVGYVLLSPFALSPDACLNTQLLRKFLCEVDKAYR-NCP 811
Query 92 YHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHP 151
YH + H + V H ++ L L L L +AAL HD+ H G+ N F+ + P
Sbjct 812 YHNSLHGSMVCHLSICLLEMLRLRESLGDLEEASLIIAALCHDIAHPGRNNNFMVNANTP 871
Query 152 LATIYNDRSILENFHASVLFR 172
LA YND S+LEN HAS+ F+
Sbjct 872 LALTYNDISVLENMHASLTFK 892
> dre:791145 pde8a, zgc:158458; phosphodiesterase 8A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=817
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 56/143 (39%), Positives = 72/143 (50%), Gaps = 11/143 (7%)
Query 38 WDADILSL-SQNYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQHIEARYDPDIPYHT 94
WD DI +L S + L +G L F LGC + LQ+IEA Y PYH
Sbjct 484 WDFDIFNLESATLKRPLAFLGLKLFSRFGVCEFLGCAEATLRSWLQNIEASYHGSNPYHN 543
Query 95 ATHAAQVAHAAMVL----NTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRH 150
+THAA V HA K LDP E +AA HDV H G+TN FL +
Sbjct 544 STHAADVLHATAYFLCKERVKQSLDPIDEVAAL----IAAAVHDVDHPGRTNSFLCNAGS 599
Query 151 PLATIYNDRSILENFHASVLFRI 173
LA +YND ++LE+ HA++ F+I
Sbjct 600 ELAVLYNDTAVLESHHAALAFQI 622
> pfa:MAL13P1.119 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase 1b (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=769
Score = 78.2 bits (191), Expect = 2e-14, Method: Composition-based stats.
Identities = 41/97 (42%), Positives = 59/97 (60%), Gaps = 1/97 (1%)
Query 78 LLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGH 137
LL ++ Y+ ++PYH + HAA V H VL + L+ L +A+L HD+GH
Sbjct 474 LLYEMKNGYN-NVPYHNSIHAAMVTHHCNVLVSNLNTANILRDNELGALFVASLGHDIGH 532
Query 138 LGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRII 174
G+TN FL+ + L+ IYND+SILEN+H S LF I+
Sbjct 533 FGRTNIFLKNCCNFLSIIYNDKSILENYHCSYLFNIL 569
> pfa:MAL13P1.118 3',5'-cyclic nucleotide phosphodiesterase (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1139
Score = 77.4 bits (189), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/97 (38%), Positives = 54/97 (55%), Gaps = 1/97 (1%)
Query 78 LLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGH 137
L +E +Y+ ++PYH HA V L KL + E + ++ + HD+GH
Sbjct 833 FLCFVEKQYN-NVPYHNTIHATMVTQKFFCLAKKLGIYDDLEYKIKLVMFISGICHDIGH 891
Query 138 LGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRII 174
G N F S HPL+ IYND S+LEN+HAS+ F+I+
Sbjct 892 PGYNNLFFVNSLHPLSIIYNDISVLENYHASITFKIL 928
> mmu:18584 Pde8a, AI551852, Pde8; phosphodiesterase 8A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=823
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 47/142 (33%), Positives = 68/142 (47%), Gaps = 3/142 (2%)
Query 38 WDADILSLSQNYQGM-LQRVGWVLLRPF--VSLLGCGNTPVIQLLQHIEARYDPDIPYHT 94
WD DI L Q L +G F L C T + Q IE+ Y PYH
Sbjct 493 WDFDIFELEVATQNRPLIYLGLKTFARFGMCEFLQCSETTLRSWFQMIESNYHSSNPYHN 552
Query 95 ATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLAT 154
+THAA V HA ++ + ++ +AA HDV H G+TN FL + + LA
Sbjct 553 STHAADVLHATAYFLSRDKIKETLDRIDEVAALIAATVHDVDHPGRTNSFLCNAGNQLAV 612
Query 155 IYNDRSILENFHASVLFRIINE 176
+YND ++LE+ H ++ F++ E
Sbjct 613 LYNDTAVLESHHVALAFQLTLE 634
> tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17 1.6.99.5)
Length=1670
Score = 77.0 bits (188), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 79/168 (47%), Gaps = 7/168 (4%)
Query 14 YESLGTAASQSFTSPPAFGGIGL------DWDADILSLSQNYQGMLQRVGWVLLRPFVSL 67
YE+L + S +P GI + DW+ D L +Q L VG+ LL
Sbjct 1254 YEALIKLVNASPRTPELSLGIPVSEEAARDWNFDCLRHAQLSPTPLVDVGYALLHRTSED 1313
Query 68 LGCGNTPVIQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLG 127
+ V++ L +E +Y+ +PYH H VA + L L+L +
Sbjct 1314 MRLPPDVVLRFLTAVEIQYN-HVPYHNCIHGLMVAQKMVALTEVLELSQSIGSRDRALVV 1372
Query 128 LAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIIN 175
+A L HD+GH G+ N + P+A +YND+S+LEN+H+ + F+ +
Sbjct 1373 VAGLCHDIGHPGRNNALFINALDPVAVLYNDKSVLENYHSCLTFKTLE 1420
> cpv:cgd3_2320 cGMP phosphodiesterase A4
Length=997
Score = 76.6 bits (187), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 39/83 (46%), Positives = 49/83 (59%), Gaps = 0/83 (0%)
Query 92 YHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHP 151
YH H V H A+ L+ L + +A+L HDVGH+G+T+ FL SRH
Sbjct 724 YHNELHGTNVCHLAICLSRATGLWSHLDTVERLASVIASLGHDVGHIGRTSNFLVNSRHM 783
Query 152 LATIYNDRSILENFHASVLFRII 174
LA YNDRS+LE FHAS+ FRII
Sbjct 784 LAINYNDRSVLEMFHASLTFRII 806
> hsa:5151 PDE8A, FLJ16150, HsT19550; phosphodiesterase 8A (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=829
Score = 76.6 bits (187), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 69/139 (49%), Gaps = 3/139 (2%)
Query 38 WDADILSL-SQNYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQHIEARYDPDIPYHT 94
WD DI L + + L +G + F L C + + LQ IEA Y PYH
Sbjct 498 WDFDIFELEAATHNRPLIYLGLKMFARFGICEFLHCSESTLRSWLQIIEANYHSSNPYHN 557
Query 95 ATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLAT 154
+TH+A V HA +K + + +AA HDV H G+TN FL + LA
Sbjct 558 STHSADVLHATAYFLSKERIKETLDPIDEVAALIAATIHDVDHPGRTNSFLCNAGSELAI 617
Query 155 IYNDRSILENFHASVLFRI 173
+YND ++LE+ HA++ F++
Sbjct 618 LYNDTAVLESHHAALAFQL 636
> dre:571011 IBMX-insensitive phosphodiesterase 8B-like; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=784
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/152 (31%), Positives = 73/152 (48%), Gaps = 6/152 (3%)
Query 28 PPAFGGI---GLDWDADILSL-SQNYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQH 81
PP+ + W+ +IL L + ++ L +G + F L C + + LQ
Sbjct 437 PPSIAELLNEEEQWEFNILELEAATHKRPLSYLGLKIFSAFGVCEFLNCSESTLRLWLQV 496
Query 82 IEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQT 141
IE Y YH +THAA V HA K + ++ LAA HDV H G+T
Sbjct 497 IETNYHSSNSYHNSTHAADVLHATAYFLRKERVKASLDQLDEVAALLAATVHDVDHPGRT 556
Query 142 NGFLQQSRHPLATIYNDRSILENFHASVLFRI 173
N FL + LA +YND ++LE+ HA++ F++
Sbjct 557 NSFLCNAGSELAILYNDTAVLESHHAALAFQL 588
> tgo:TGME49_020420 3'5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=466
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/141 (34%), Positives = 68/141 (48%), Gaps = 23/141 (16%)
Query 34 IGLDWDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYH 93
+G DWD D+ L + M +L+ G +L Q+ R++P YH
Sbjct 122 VGEDWDLDMFELCKQTNRM-------------TLMATG----FRLQQY---RHNP---YH 158
Query 94 TATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLA 153
H A VAH + L + + +AAL HDVGH N ++ S HPLA
Sbjct 159 NEQHGAAVAHMMVFLLRACQAWHMFKPLYQTAIIVAALVHDVGHFAMNNHYVVNSGHPLA 218
Query 154 TIYNDRSILENFHASVLFRII 174
YNDRS+LENFH+++ FRI+
Sbjct 219 ITYNDRSVLENFHSALAFRIM 239
> mmu:218461 Pde8b, B230331L10Rik, C030047E14Rik; phosphodiesterase
8B (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=788
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 50/153 (32%), Positives = 73/153 (47%), Gaps = 8/153 (5%)
Query 28 PPAFGGIGLD----WDADILSLSQ-NYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQ 80
PP+ + LD WD +I L ++ L +G + F L C T + LQ
Sbjct 447 PPSIAQL-LDNEESWDFNIFELEAVTHKRPLVYLGLKVFSRFGVCEFLNCTETTLRAWLQ 505
Query 81 HIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQ 140
IEA Y YH +THAA V HA K + ++ +AA HDV H G+
Sbjct 506 VIEANYHSSNAYHNSTHAADVLHATAFFLGKERVKGSLDQLDEVAALIAATVHDVDHPGR 565
Query 141 TNGFLQQSRHPLATIYNDRSILENFHASVLFRI 173
TN FL + LA +YND ++LE+ H ++ F++
Sbjct 566 TNSFLCNAGSELAVLYNDTAVLESHHTALAFQL 598
> tpv:TP01_1200 hypothetical protein
Length=840
Score = 71.6 bits (174), Expect = 1e-12, Method: Composition-based stats.
Identities = 43/141 (30%), Positives = 67/141 (47%), Gaps = 3/141 (2%)
Query 37 DWDADILS-LSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEARYDPDIPYHTA 95
DW+ IL QN G + +G VLL F + + L +E Y+ ++ YH
Sbjct 552 DWNFSILDYFRQNPTGFIS-IGCVLLSEFQTHFNIPTDVIYSFLGLVEKCYN-NVSYHNQ 609
Query 96 THAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLATI 155
H V + L+ ++ L ++ L HD+GH G TN F S H LA +
Sbjct 610 MHGVFVCQKLLCLSNFTNVYSRLSVIDRTILIISGLCHDIGHPGLTNAFFINSGHQLAHL 669
Query 156 YNDRSILENFHASVLFRIINE 176
+ND+S+LENFH + F ++ +
Sbjct 670 FNDKSVLENFHCCLTFNVLRQ 690
> hsa:8622 PDE8B, ADSD, FLJ11212, PPNAD3; phosphodiesterase 8B
(EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=788
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/153 (32%), Positives = 71/153 (46%), Gaps = 8/153 (5%)
Query 28 PPAFGGIGLD----WDADILSLSQ-NYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQ 80
PP + LD WD +I L ++ L +G + F L C T + Q
Sbjct 447 PPCISQL-LDNEESWDFNIFELEAITHKRPLVYLGLKVFSRFGVCEFLNCSETTLRAWFQ 505
Query 81 HIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQ 140
IEA Y YH +THAA V HA K + ++ +AA HDV H G+
Sbjct 506 VIEANYHSSNAYHNSTHAADVLHATAFFLGKERVKGSLDQLDEVAALIAATVHDVDHPGR 565
Query 141 TNGFLQQSRHPLATIYNDRSILENFHASVLFRI 173
TN FL + LA +YND ++LE+ H ++ F++
Sbjct 566 TNSFLCNAGSELAVLYNDTAVLESHHTALAFQL 598
> cpv:cgd6_500 membrane associated HD superfamily cyclic nucleotide
phosphodiesterase domain containing protein
Length=1967
Score = 71.2 bits (173), Expect = 2e-12, Method: Composition-based stats.
Identities = 46/144 (31%), Positives = 64/144 (44%), Gaps = 10/144 (6%)
Query 43 LSLSQNYQGMLQRVGWVLLRPFV----------SLLGCGNTPVIQLLQHIEARYDPDIPY 92
LSL + + + GW + R F+ S N ++ L I Y+ DIPY
Sbjct 1492 LSLYREWGFPIWAHGWSVHREFLLNLFKYYGFDSKWNWSNKQLLTLFDLIHDSYNSDIPY 1551
Query 93 HTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPL 152
H HA QV ++ + F L AAL HD+ H G N FL S L
Sbjct 1552 HNIFHALQVVQVCYIILRNFGIMLVLNDLNKFILLFAALCHDIDHPGVNNCFLTISNSKL 1611
Query 153 ATIYNDRSILENFHASVLFRIINE 176
A YND SILEN H + +F ++++
Sbjct 1612 ALKYNDNSILENHHCNYIFSLLDK 1635
> hsa:5142 PDE4B, DKFZp686F2182, DPDE4, MGC126529, PDE4B5, PDEIVB;
phosphodiesterase 4B, cAMP-specific (EC:3.1.4.17); K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=564
Score = 65.9 bits (159), Expect = 7e-11, Method: Composition-based stats.
Identities = 38/101 (37%), Positives = 52/101 (51%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
I + +E Y D+ YH + HAA VA + VL + LD AA HDV
Sbjct 217 ITYMMTLEDHYHSDVAYHNSLHAADVAQSTHVLLSTPALDAVFTDLEILAAIFAAAIHDV 276
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 277 DHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQE 317
> mmu:18578 Pde4b, Dpde4, R74983, dunce; phosphodiesterase 4B,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=564
Score = 65.5 bits (158), Expect = 1e-10, Method: Composition-based stats.
Identities = 37/101 (36%), Positives = 52/101 (51%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
+ + +E Y D+ YH + HAA VA + VL + LD AA HDV
Sbjct 217 VTYMMTLEDHYHSDVAYHNSLHAADVAQSTHVLLSTPALDAVFTDLEILAAIFAAAIHDV 276
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 277 DHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQE 317
> dre:565259 pde4c, im:7160317, si:dkey-149i17.5; phosphodiesterase
4C, cAMP-specific (phosphodiesterase E1 dunce homolog,
Drosophila)
Length=672
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 35/101 (34%), Positives = 51/101 (50%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
I + +E Y D+ YH + HAA V + VL + L+ A+ HDV
Sbjct 357 INYMTALEENYRSDVAYHNSIHAADVVQSTHVLLSTPALEAVFTDLEILAAMFASAIHDV 416
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 417 DHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQE 457
> mmu:238871 Pde4d, 9630011N22Rik, Dpde3; phosphodiesterase 4D,
cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=747
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 36/102 (35%), Positives = 51/102 (50%), Gaps = 0/102 (0%)
Query 75 VIQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHD 134
+I L +E Y D+ YH HAA V + VL + L+ A+ HD
Sbjct 383 LITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPALEAVFTDLEILAAIFASAIHD 442
Query 135 VGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
V H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 443 VDHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQE 484
> mmu:18575 Pde1c; phosphodiesterase 1C (EC:3.1.4.17); K13755
calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=631
Score = 64.7 bits (156), Expect = 2e-10, Method: Composition-based stats.
Identities = 43/143 (30%), Positives = 68/143 (47%), Gaps = 4/143 (2%)
Query 38 WDADILSLSQ-NYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQHIEARYDPDI-PYH 93
W D+ SL++ + L+ + + LL + +S + ++ ++ +E Y PYH
Sbjct 169 WSFDVFSLNEASGDHALKFIFYELLTRYDLISRFKIPISALVSFVEALEVGYSKHKNPYH 228
Query 94 TATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLA 153
HAA V L K + + F + +A HD H G TN F Q+R A
Sbjct 229 NLMHAADVTQTVHYLLYKTGVANWLTELEIFAIIFSAAIHDYEHTGTTNNFHIQTRSDPA 288
Query 154 TIYNDRSILENFHASVLFRIINE 176
+YNDRS+LEN H S +R++ E
Sbjct 289 ILYNDRSVLENHHLSAAYRLLQE 311
> xla:495146 hypothetical LOC495146; K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=682
Score = 64.7 bits (156), Expect = 2e-10, Method: Composition-based stats.
Identities = 41/113 (36%), Positives = 56/113 (49%), Gaps = 3/113 (2%)
Query 67 LLGCGNTPVIQLLQH---IEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGS 123
LL PV L+ + +E Y D+ YH + HAA V + VL + LD
Sbjct 315 LLKTFQIPVDTLMTYMMTLEDHYHADVAYHNSLHAADVTQSTHVLLSTPALDAVFTDLEI 374
Query 124 FCLGLAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
AA HDV H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 375 LAALFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQE 427
> dre:566998 novel protein similar to vertebrate phosphodiesterase
4D, cAMP-specific (phosphodiesterase E3 dunce homolog,
Drosophila) (PDE4D); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=745
Score = 64.3 bits (155), Expect = 2e-10, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 50/101 (49%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
I L +E Y D+ YH HAA V + VL + L+ A+ HDV
Sbjct 357 ITYLMTLEDHYHADVAYHNNIHAADVTQSTHVLLSTPALEAVFTDLEILAAIFASAIHDV 416
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 417 DHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQE 457
> dre:571344 fc68c05, si:ch211-255d18.8, wu:fc68c05; si:ch211-255d18.1
Length=709
Score = 64.3 bits (155), Expect = 2e-10, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 50/101 (49%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
+ L +E Y D+ YH HAA V + VL + L+ A+ HDV
Sbjct 343 LTFLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPALEDVFTDLEIMAALFASAIHDV 402
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G TN FL + LA +YND S+LEN H +V F+++ E
Sbjct 403 DHPGVTNQFLINTNSELALMYNDASVLENHHLAVGFKLLQE 443
> tgo:TGME49_093000 3', 5'-cyclic nucleotide phosphodiesterase
domain-containing protein (EC:3.2.1.3 3.1.4.17 3.2.1.108)
Length=2098
Score = 64.3 bits (155), Expect = 2e-10, Method: Composition-based stats.
Identities = 34/92 (36%), Positives = 46/92 (50%), Gaps = 0/92 (0%)
Query 86 YDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFL 145
Y D PYH HA V + + + LAAL HDV H G N FL
Sbjct 1757 YHTDNPYHNFYHAIHVFQVCWMFLSAYGCRNILSPIEQLGILLAALCHDVDHPGVNNAFL 1816
Query 146 QQSRHPLATIYNDRSILENFHASVLFRIINEI 177
+ S HPL+ +YND+S+LEN HA+ R + ++
Sbjct 1817 RASLHPLSIMYNDKSVLENHHAAFAIRTMMQL 1848
> dre:565706 cAMP-specific 3,5-cyclic phosphodiesterase 4B-like;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=713
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 52/101 (51%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
+ + +E Y D+ YH + HAA VA + +L + LD AA HDV
Sbjct 367 VTYMMTLEDHYHQDVAYHNSLHAADVAQSTHILLSTPALDAVFTDLEILAAIFAAAIHDV 426
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 427 DHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQE 467
> xla:444562 pde4b, MGC83972; phosphodiesterase 4B, cAMP-specific;
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=721
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 41/113 (36%), Positives = 56/113 (49%), Gaps = 3/113 (2%)
Query 67 LLGCGNTPVIQLLQH---IEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGS 123
LL PV L+ + +E Y D+ YH + HAA V + VL + LD
Sbjct 363 LLKTFKIPVDTLITYTMTLEDHYHSDVAYHNSLHAADVTQSTHVLLSTPALDAVFTDLEI 422
Query 124 FCLGLAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
AA HDV H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 423 LAAIFAAAIHDVDHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLQE 475
> tgo:TGME49_033070 phosphodiesterase, putative (EC:3.1.4.17)
Length=397
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 49/84 (58%), Gaps = 0/84 (0%)
Query 91 PYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRH 150
PYH HA VA +L D+ + F L +AAL HD+GH G N F+ ++
Sbjct 79 PYHNFFHALNVAQVCCLLMALPDVAARFQPLDYFVLSVAALGHDLGHPGANNLFVNRNDC 138
Query 151 PLATIYNDRSILENFHASVLFRII 174
+ +Y +RS+LEN+HA++LF+I+
Sbjct 139 LPSRLYQNRSVLENYHAALLFQIL 162
> hsa:5137 PDE1C, Hcam3; phosphodiesterase 1C, calmodulin-dependent
70kDa (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=634
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 42/143 (29%), Positives = 68/143 (47%), Gaps = 4/143 (2%)
Query 38 WDADILSLSQ-NYQGMLQRVGWVLLRPF--VSLLGCGNTPVIQLLQHIEARYDPDI-PYH 93
W D+ SL++ + L+ + + LL + +S + ++ ++ +E Y PYH
Sbjct 169 WSFDVFSLNEASGDHALKFIFYELLTRYDLISRFKIPISALVSFVEALEVGYSKHKNPYH 228
Query 94 TATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLA 153
HAA V L K + + F + +A HD H G TN F Q+R A
Sbjct 229 NLMHAADVTQTVHYLLYKTGVANWLTELEIFAIIFSAAIHDYEHTGTTNNFHIQTRSDPA 288
Query 154 TIYNDRSILENFHASVLFRIINE 176
+YNDRS+LEN H S +R++ +
Sbjct 289 ILYNDRSVLENHHLSAAYRLLQD 311
> cel:T04D3.3 pde-1; PhosphoDiEsterase family member (pde-1);
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=664
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 45/143 (31%), Positives = 65/143 (45%), Gaps = 4/143 (2%)
Query 38 WDADILSLSQNYQG-MLQRVGWVLLRP--FVSLLGCGNTPVIQLLQHIEARYDP-DIPYH 93
W L++ +G L+ VG+ L F+ T + L +E Y + PYH
Sbjct 274 WSFSPFQLNEVSEGHALKYVGFELFNRYGFMDRFKVPLTALENYLSALEVGYSKHNNPYH 333
Query 94 TATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLA 153
HAA V ++ + ++ L + AL HD H G TN F QS+ A
Sbjct 334 NVVHAADVTQSSHFMLSQTGLANSLGDLELLAVLFGALIHDYEHTGHTNNFHIQSQSQFA 393
Query 154 TIYNDRSILENFHASVLFRIINE 176
+YNDRS+LEN H S FR++ E
Sbjct 394 MLYNDRSVLENHHVSSCFRLMKE 416
> dre:564465 pde1a; phosphodiesterase 1A, calmodulin-dependent;
K13755 calcium/calmodulin-dependent 3',5'-cyclic nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=625
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 49/160 (30%), Positives = 70/160 (43%), Gaps = 9/160 (5%)
Query 25 FTSPPAFGGIGLD---WDADILSLSQ-NYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQ 80
T PP+ D W D+ L + + L+ + + LL + L+ PV L+
Sbjct 147 LTYPPSVITALKDVDKWSFDVFKLQEASSDHALKFLVYELLTRY-DLISRFRIPVSSLVS 205
Query 81 HIEA----RYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVG 136
+EA PYH HAA V A L + + + AA HD
Sbjct 206 FVEALEIGYSKHRNPYHNLIHAADVTQTAHYLMLHTGVMHWLTELEILAMVFAAAIHDFE 265
Query 137 HLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G TN F Q+R +A +YNDRS+LEN H S +R++ E
Sbjct 266 HTGTTNNFHIQTRSEVAILYNDRSVLENHHVSAAYRLMQE 305
> mmu:18574 Pde1b, 63kDa, Pde1b1; phosphodiesterase 1B, Ca2+-calmodulin
dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=535
Score = 62.0 bits (149), Expect = 1e-09, Method: Composition-based stats.
Identities = 43/145 (29%), Positives = 63/145 (43%), Gaps = 8/145 (5%)
Query 38 WDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEA------RYDPDIP 91
W D+ SL++ R L SL+ P + L+ +EA +Y P
Sbjct 163 WCFDVFSLNRAADDHALRTIVFELLTRHSLISRFKIPTVFLMSFLEALETGYGKYKN--P 220
Query 92 YHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHP 151
YH HAA V + + + + AA HD H G TN F Q++
Sbjct 221 YHNQIHAADVTQTVHCFLLRTGMVHCLSEIEVLAIIFAAAIHDYEHTGTTNSFHIQTKSE 280
Query 152 LATIYNDRSILENFHASVLFRIINE 176
A +YNDRS+LEN H S +FR++ +
Sbjct 281 CAILYNDRSVLENHHISSVFRMMQD 305
> dre:572061 pde4a; phosphodiesterase 4A, cAMP-specific; K01120
3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=736
Score = 61.6 bits (148), Expect = 1e-09, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 52/101 (51%), Gaps = 0/101 (0%)
Query 76 IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDV 135
I + +E Y ++ YH + HAA V + VL + LD AA HDV
Sbjct 375 ITYVMTLEDHYHANVAYHNSLHAADVTQSTHVLLSTPALDAVFSDLEILAALFAAAIHDV 434
Query 136 GHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
H G +N FL + LA +YND S+LEN H +V F++++E
Sbjct 435 DHPGVSNQFLINTNSELALMYNDESVLENHHLAVGFKLLHE 475
> hsa:5136 PDE1A, HCAM1, HSPDE1A, MGC26303; phosphodiesterase
1A, calmodulin-dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=535
Score = 61.6 bits (148), Expect = 1e-09, Method: Composition-based stats.
Identities = 31/86 (36%), Positives = 42/86 (48%), Gaps = 0/86 (0%)
Query 91 PYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRH 150
PYH HAA V + + + + AA HD H G TN F Q+R
Sbjct 217 PYHNLIHAADVTQTVHYIMLHTGIMHWLTELEILAMVFAAAIHDYEHTGTTNNFHIQTRS 276
Query 151 PLATIYNDRSILENFHASVLFRIINE 176
+A +YNDRS+LEN H S +R++ E
Sbjct 277 DVAILYNDRSVLENHHVSAAYRLMQE 302
> hsa:5153 PDE1B, PDE1B1, PDES1B; phosphodiesterase 1B, calmodulin-dependent
(EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase [EC:3.1.4.17]
Length=536
Score = 61.2 bits (147), Expect = 2e-09, Method: Composition-based stats.
Identities = 42/145 (28%), Positives = 63/145 (43%), Gaps = 8/145 (5%)
Query 38 WDADILSLSQNYQGMLQRVGWVLLRPFVSLLGCGNTPVIQLLQHIEA------RYDPDIP 91
W D+ SL+Q R L +L+ P + L+ ++A +Y P
Sbjct 164 WCFDVFSLNQAADDHALRTIVFELLTRHNLISRFKIPTVFLMSFLDALETGYGKYKN--P 221
Query 92 YHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHP 151
YH HAA V + + + + AA HD H G TN F Q++
Sbjct 222 YHNQIHAADVTQTVHCFLLRTGMVHCLSEIELLAIIFAAAIHDYEHTGTTNSFHIQTKSE 281
Query 152 LATIYNDRSILENFHASVLFRIINE 176
A +YNDRS+LEN H S +FR++ +
Sbjct 282 CAIVYNDRSVLENHHISSVFRLMQD 306
> hsa:5144 PDE4D, DKFZp686M11213, DPDE3, FLJ97311, HSPDE4D, PDE4DN2,
STRK1; phosphodiesterase 4D, cAMP-specific (EC:3.1.4.17);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=809
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 54/113 (47%), Gaps = 3/113 (2%)
Query 67 LLGCGNTPV---IQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGS 123
LL PV I L +E Y D+ YH HAA V + VL + L+
Sbjct 433 LLKTFKIPVDTLITYLMTLEDHYHADVAYHNNIHAADVVQSTHVLLSTPALEAVFTDLEI 492
Query 124 FCLGLAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIINE 176
A+ HDV H G +N FL + LA +YND S+LEN H +V F+++ E
Sbjct 493 LAAIFASAIHDVDHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQE 545
> dre:567051 si:dkey-48h7.2; K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1117
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 56/186 (30%), Positives = 74/186 (39%), Gaps = 51/186 (27%)
Query 37 DWDADILSLSQNYQGMLQRVGWVL----LRPFVS--LLGCGNTPVIQLLQHIEARYD--P 88
+W+ I SL + G R G +L R FV L PV + + + A +
Sbjct 621 NWNFPIFSLVEKTNG---RCGCILSQMSYRLFVDTGLFETFKIPVREFMNYFHALENGYR 677
Query 89 DIPYHTATHAAQVAHAAMVLNTK-------------LDLDPG---------------KEK 120
DIPYH HA V HA L T+ D D G E+
Sbjct 678 DIPYHNRIHATDVLHAVWYLTTQPVPGLPTLRTLGSSDSDSGLAHGRMKYLMSRTNAVEE 737
Query 121 TGSFCLG------------LAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHAS 168
G CL +AA HD H G+TN FL + P A +YNDRS+LEN HA+
Sbjct 738 EGYGCLSGLIPALELMALYVAAAMHDYDHPGRTNAFLVATSAPQAVLYNDRSVLENHHAA 797
Query 169 VLFRII 174
+ +
Sbjct 798 SAWNLF 803
> cel:Y95B8A.10 pde-6; PhosphoDiEsterase family member (pde-6);
K01120 3',5'-cyclic-nucleotide phosphodiesterase [EC:3.1.4.17]
Length=760
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 54/178 (30%), Positives = 81/178 (45%), Gaps = 7/178 (3%)
Query 1 RGKNRSAVHGKGSYESLGTAASQSFTSPPAFGGIGLD--WDADILSLSQ-NYQGMLQRVG 57
R + RS V E G+ + S + D W DIL L + + L +VG
Sbjct 407 RQRKRSVV--DAHREKRGSHGERRRVSADVKNALENDNCWKFDILHLEKVSDHHALSQVG 464
Query 58 WVLLRPF--VSLLGCGNTPVIQLLQHIEARYDPDIPYHTATHAAQVAHAAMVLNTKLDLD 115
+ + +LGC + + + + IEA Y YH ATHAA V A +
Sbjct 465 MKVFERWKVCDVLGCSDDLLHRWILSIEAHYHAGNTYHNATHAADVLQATSFFLDSPSVA 524
Query 116 PGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRI 173
++ + LAA HD+ H G+ N +L +R LA +YND SILEN H ++ F++
Sbjct 525 VHVNESHAVAALLAAAVHDLDHPGRGNAYLINTRQSLAILYNDNSILENHHIALAFQL 582
> mmu:18573 Pde1a, AI987702, AW125737, MGC116577; phosphodiesterase
1A, calmodulin-dependent (EC:3.1.4.17); K13755 calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=545
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 42/86 (48%), Gaps = 0/86 (0%)
Query 91 PYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGSFCLGLAALAHDVGHLGQTNGFLQQSRH 150
PYH HAA V + + + + AA HD H G TN F Q+R
Sbjct 217 PYHNLVHAADVTQTVHYIMLHTGIMHWLTELEILAMVFAAAIHDYEHTGTTNNFHIQTRS 276
Query 151 PLATIYNDRSILENFHASVLFRIINE 176
+A +YNDRS+LEN H S +R++ E
Sbjct 277 DVAILYNDRSVLENHHVSAAYRLMQE 302
> mmu:110385 Pde4c, Dpde1, E130301F19Rik, MGC31320, dunce; phosphodiesterase
4C, cAMP specific (EC:3.1.4.17); K01120 3',5'-cyclic-nucleotide
phosphodiesterase [EC:3.1.4.17]
Length=652
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 53/112 (47%), Gaps = 3/112 (2%)
Query 67 LLGCGNTPVIQLLQHI---EARYDPDIPYHTATHAAQVAHAAMVLNTKLDLDPGKEKTGS 123
LL P LL ++ E Y D+ YH + HAA V +A VL L+
Sbjct 360 LLKTFQIPADTLLAYLLTLEGHYHSDVAYHNSMHAADVVQSAHVLLGTPALEAVFTDLEV 419
Query 124 FCLGLAALAHDVGHLGQTNGFLQQSRHPLATIYNDRSILENFHASVLFRIIN 175
A HDV H G +N FL + LA +YND S+LEN H +V F+++
Sbjct 420 LAAIFACAIHDVDHPGVSNQFLINTNSELALMYNDSSVLENHHLAVGFKLLQ 471
Lambda K H
0.320 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40