bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1494_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
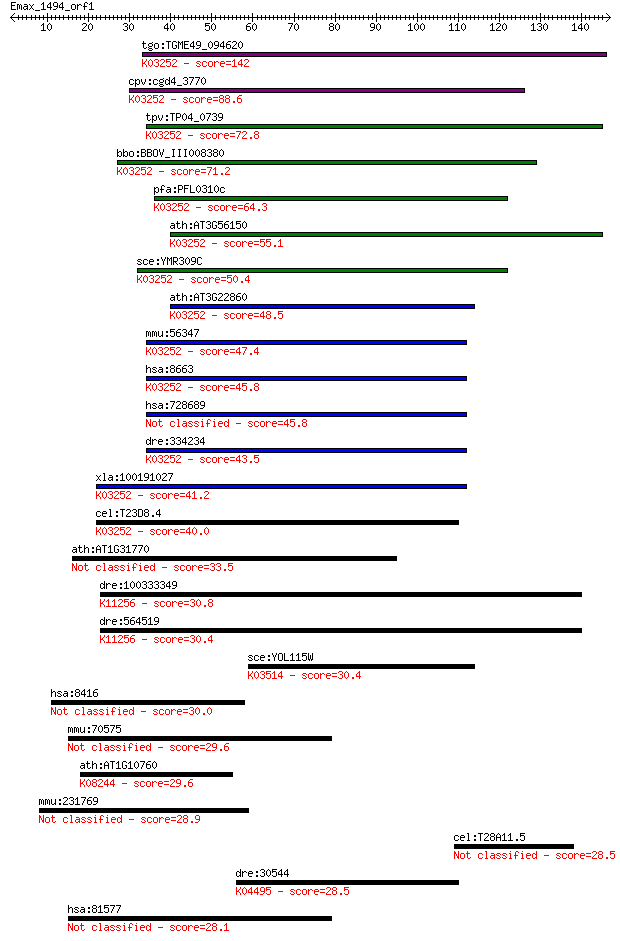
tgo:TGME49_094620 eukaryotic translation initiation factor 3 s... 142 4e-34
cpv:cgd4_3770 hypothetical protein ; K03252 translation initia... 88.6 6e-18
tpv:TP04_0739 eukaryotic translation initiation factor 3 subun... 72.8 4e-13
bbo:BBOV_III008380 17.m07733; eukaryotic translation initiatio... 71.2 1e-12
pfa:PFL0310c eukaryotic translation initiation factor 3 subuni... 64.3 1e-10
ath:AT3G56150 EIF3C; EIF3C (EUKARYOTIC TRANSLATION INITIATION ... 55.1 7e-08
sce:YMR309C NIP1; Nip1p; K03252 translation initiation factor ... 50.4 2e-06
ath:AT3G22860 TIF3C2; TIF3C2; translation initiation factor; K... 48.5 6e-06
mmu:56347 Eif3c, 110kDa, 3230401O13Rik, Eif3s8, MGC36637, NIPI... 47.4 1e-05
hsa:8663 EIF3C, EIF3CL, EIF3S8, FLJ53378, FLJ54400, FLJ54404, ... 45.8 4e-05
hsa:728689 EIF3CL; eukaryotic translation initiation factor 3,... 45.8 4e-05
dre:334234 eif3c, eif3s8, fi37c04, wu:fb34a08, wu:fi37c04, zgc... 43.5 2e-04
xla:100191027 eif3c, MGC114671, eif3-p110, eif3s8; eukaryotic ... 41.2 0.001
cel:T23D8.4 eif-3.C; Eukaryotic Initiation Factor family membe... 40.0 0.002
ath:AT1G31770 ABC transporter family protein 33.5 0.23
dre:100333349 nuclear receptor coactivator 2-like; K11256 nucl... 30.8 1.6
dre:564519 ncoa3, sb:eu248, sb:eu484, src3; nuclear receptor c... 30.4 2.1
sce:YOL115W PAP2, TRF4; Non-canonical poly(A) polymerase, invo... 30.4 2.3
hsa:8416 ANXA9, ANX31; annexin A9 30.0 2.7
mmu:70575 Gfod2, 5730466C23Rik; glucose-fructose oxidoreductas... 29.6 3.4
ath:AT1G10760 SEX1; SEX1 (STARCH EXCESS 1); alpha-glucan, wate... 29.6 3.6
mmu:231769 Sfswap, 1190005N23Rik, 6330437E22Rik, AI197402, AW2... 28.9 5.9
cel:T28A11.5 hypothetical protein 28.5 7.6
dre:30544 sox17, MGC91776; SRY-box containing gene 17; K04495 ... 28.5 7.8
hsa:81577 GFOD2, FLJ23802, MGC11335; glucose-fructose oxidored... 28.1 9.7
> tgo:TGME49_094620 eukaryotic translation initiation factor 3
subunit 8, putative ; K03252 translation initiation factor
3 subunit C
Length=1104
Score = 142 bits (358), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 66/113 (58%), Positives = 88/113 (77%), Gaps = 0/113 (0%)
Query 33 QQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFK 92
Q +S+ ILTSFVERLDD+LMKSLQ DVHS+EYKERLG+S+D++A+L R + L + K
Sbjct 438 QVVSSGILTSFVERLDDELMKSLQFTDVHSEEYKERLGQSVDMIAVLCRAWVHLTDCQSK 497
Query 93 TQAATVALKVNEHMHYKPDNIAIKMWDVLRKELPEELCKDLPGENIPPTAFIE 145
QAAT+AL++NEHMHYK D IA MWD++RK +P E+ + LP E + P+ F+E
Sbjct 498 PQAATLALRINEHMHYKHDAIAASMWDLVRKRVPAEVAQHLPDEGMKPSDFVE 550
> cpv:cgd4_3770 hypothetical protein ; K03252 translation initiation
factor 3 subunit C
Length=1040
Score = 88.6 bits (218), Expect = 6e-18, Method: Composition-based stats.
Identities = 39/96 (40%), Positives = 68/96 (70%), Gaps = 0/96 (0%)
Query 30 EEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDER 89
+E + + + L SF+ERLD + +K+LQ DVHS EYK+RL +S+ +LALLWR + +ER
Sbjct 472 KEQDKSTVTFLVSFIERLDGESLKALQLTDVHSSEYKDRLVQSLHLLALLWRCYKICEER 531
Query 90 GFKTQAATVALKVNEHMHYKPDNIAIKMWDVLRKEL 125
G+ +++++ + +H+K D++AIK+W+ +R+ L
Sbjct 532 GYYDLVSSLSVHLINQLHFKNDSLAIKVWEFVRQIL 567
> tpv:TP04_0739 eukaryotic translation initiation factor 3 subunit
8; K03252 translation initiation factor 3 subunit C
Length=951
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 71/115 (61%), Gaps = 5/115 (4%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFL--DERGF 91
+ S ++L++ V++L+D+L K L +VH+ +YK L +ID+L LL RT + E+G
Sbjct 459 KTSLTVLSTIVQKLNDELYKGLLYTEVHNPDYKTMLAYTIDMLYLLHRTLVYYLKFEKGN 518
Query 92 KTQAATVALKVNEHMHYKPDNIAIKMWDVLRKELPE--ELCKDLPGENIPPTAFI 144
+ AAT AL + +H HYK D I+ K+W+++R ++ + + + P EN P+ +
Sbjct 519 EF-AATTALMILDHCHYKDDEISTKIWELVRNKIKDKKQTERFFPNENKKPSDLV 572
> bbo:BBOV_III008380 17.m07733; eukaryotic translation initiation
factor 3 subunit 8; K03252 translation initiation factor
3 subunit C
Length=966
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 69/104 (66%), Gaps = 3/104 (2%)
Query 27 TTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFL 86
+ EE IQ+ S +IL S V +L+D+L K L I+V SD+Y L ++++L LL +T +
Sbjct 474 SREERIQR-SMTILESVVRKLNDELYKGLLYIEVGSDDYNTMLVYNVNMLYLLHKTLLYC 532
Query 87 DERGFK--TQAATVALKVNEHMHYKPDNIAIKMWDVLRKELPEE 128
+G + T AA +A+ + EH+HYK D ++ K+W+++R+++ +E
Sbjct 533 LSKGKETITHAANIAIIMLEHLHYKDDVVSGKIWELVRQKVTDE 576
> pfa:PFL0310c eukaryotic translation initiation factor 3 subunit
8, putative; K03252 translation initiation factor 3 subunit
C
Length=984
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 31/87 (35%), Positives = 57/87 (65%), Gaps = 1/87 (1%)
Query 36 SASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLD-ERGFKTQ 94
S L SF+ +LDD+L+K+L IDV ++EY++RLGK+I ++ LL + + ++ +
Sbjct 464 SCKTLISFLAKLDDELLKALLYIDVQTEEYRKRLGKTIHMIGLLKKGYNYVKCLKNMPDL 523
Query 95 AATVALKVNEHMHYKPDNIAIKMWDVL 121
A ++ ++ EHM+YKP+ + ++W L
Sbjct 524 AIHISSRILEHMYYKPEMLFKQIWTYL 550
> ath:AT3G56150 EIF3C; EIF3C (EUKARYOTIC TRANSLATION INITIATION
FACTOR 3C); translation initiation factor; K03252 translation
initiation factor 3 subunit C
Length=900
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 39/105 (37%), Positives = 55/105 (52%), Gaps = 0/105 (0%)
Query 40 LTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKTQAATVA 99
L +F+ER+D + KSLQ ID H+ EY ERL LAL + + G AA VA
Sbjct 356 LVAFLERVDTEFFKSLQCIDPHTREYVERLRDEPMFLALAQNIQDYFERMGDFKAAAKVA 415
Query 100 LKVNEHMHYKPDNIAIKMWDVLRKELPEELCKDLPGENIPPTAFI 144
L+ E ++YKP + M + EE ++ E+ PPT+FI
Sbjct 416 LRRVEAIYYKPQEVYDAMRKLAELVEEEEETEEAKEESGPPTSFI 460
> sce:YMR309C NIP1; Nip1p; K03252 translation initiation factor
3 subunit C
Length=812
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 53/99 (53%), Gaps = 10/99 (10%)
Query 32 IQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFL----- 86
+++I SI SFVERLDD+ MKSL +ID HS +Y RL I L+ RT +
Sbjct 362 VKRILGSIF-SFVERLDDEFMKSLLNIDPHSSDYLIRLRDEQSIYNLILRTQLYFEATLK 420
Query 87 DERGFKTQAATVALKVNEHMHYKPDNIAIKM----WDVL 121
DE + +K +H++YK +N+ M W+++
Sbjct 421 DEHDLERALTRPFVKRLDHIYYKSENLIKIMETAAWNII 459
> ath:AT3G22860 TIF3C2; TIF3C2; translation initiation factor;
K03252 translation initiation factor 3 subunit C
Length=805
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 42/74 (56%), Gaps = 0/74 (0%)
Query 40 LTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKTQAATVA 99
L +F+E+++ + KSLQ ID H+++Y ERL LAL +L+ G A+ VA
Sbjct 318 LVAFLEKIETEFFKSLQCIDPHTNDYVERLKDEPMFLALAQSIQDYLERTGDSKAASKVA 377
Query 100 LKVNEHMHYKPDNI 113
+ E ++YKP +
Sbjct 378 FILVESIYYKPQEV 391
> mmu:56347 Eif3c, 110kDa, 3230401O13Rik, Eif3s8, MGC36637, NIPIL(A3),
NipilA3; eukaryotic translation initiation factor 3,
subunit C; K03252 translation initiation factor 3 subunit
C
Length=911
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 43/78 (55%), Gaps = 1/78 (1%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKT 93
++ ILT VER+D++ K +Q+ D HS EY E L + A++ R +L+E+G
Sbjct 437 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 495
Query 94 QAATVALKVNEHMHYKPD 111
+ + L+ H +YK D
Sbjct 496 EICQIYLRRILHTYYKFD 513
> hsa:8663 EIF3C, EIF3CL, EIF3S8, FLJ53378, FLJ54400, FLJ54404,
FLJ55450, FLJ55750, FLJ78287, MGC189737, MGC189744, eIF3-p110;
eukaryotic translation initiation factor 3, subunit C;
K03252 translation initiation factor 3 subunit C
Length=913
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKT 93
++ ILT VER+D++ K +Q+ D HS EY E L + A++ R +L+E+G
Sbjct 439 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 497
Query 94 QAATVALKVNEHMHYKPD 111
+ + L H +YK D
Sbjct 498 EVCRIYLLRILHTYYKFD 515
> hsa:728689 EIF3CL; eukaryotic translation initiation factor
3, subunit C-like
Length=914
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKT 93
++ ILT VER+D++ K +Q+ D HS EY E L + A++ R +L+E+G
Sbjct 440 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVEHLKDEAQVCAIIERVQRYLEEKGTTE 498
Query 94 QAATVALKVNEHMHYKPD 111
+ + L H +YK D
Sbjct 499 EVCRIYLLRILHTYYKFD 516
> dre:334234 eif3c, eif3s8, fi37c04, wu:fb34a08, wu:fi37c04, zgc:66128;
eukaryotic translation initiation factor 3, subunit
C; K03252 translation initiation factor 3 subunit C
Length=926
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 41/78 (52%), Gaps = 1/78 (1%)
Query 34 QISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKT 93
++ ILT VER+D++ K +Q+ D HS EY + L + ++ R +L+ +G
Sbjct 439 RVRGCILT-LVERMDEEFTKIMQNTDPHSQEYVDNLKDEGRVCGIIDRLLQYLETKGSTE 497
Query 94 QAATVALKVNEHMHYKPD 111
+ V L+ H +YK D
Sbjct 498 EVCRVYLRRIMHTYYKFD 515
> xla:100191027 eif3c, MGC114671, eif3-p110, eif3s8; eukaryotic
translation initiation factor 3, subunit C; K03252 translation
initiation factor 3 subunit C
Length=926
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 48/95 (50%), Gaps = 6/95 (6%)
Query 22 EAITETTE-----EEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDIL 76
E I E +E E+ ++ ILT +ER++++ K +Q+ D HS EY + L +
Sbjct 425 EHIVEDSENLSNLEQPLRVRGCILT-LIERMEEEFTKIMQNTDPHSQEYVDNLKDEARVC 483
Query 77 ALLWRTFCFLDERGFKTQAATVALKVNEHMHYKPD 111
++ R +L E+G + V L+ H +YK D
Sbjct 484 EVIERAQKYLQEKGSTEEICRVYLRRIMHTYYKFD 518
> cel:T23D8.4 eif-3.C; Eukaryotic Initiation Factor family member
(eif-3.C); K03252 translation initiation factor 3 subunit
C
Length=898
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 8/93 (8%)
Query 22 EAITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWR 81
E + + T+E +I SIL + V+RLD +L K LQ+ D HS++Y E+L D+ +L+ +
Sbjct 404 ENLKDDTQE--YRIQGSILIA-VQRLDGELAKILQNADCHSNDYIEKLKAEKDMCSLIEK 460
Query 82 TFCFLDERG-----FKTQAATVALKVNEHMHYK 109
+++ R K + V + EH +YK
Sbjct 461 AEKYVELRNDSGIFDKHEVCKVYMMRIEHAYYK 493
> ath:AT1G31770 ABC transporter family protein
Length=648
Score = 33.5 bits (75), Expect = 0.23, Method: Composition-based stats.
Identities = 18/82 (21%), Positives = 40/82 (48%), Gaps = 3/82 (3%)
Query 16 AQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSI-- 73
A G+ + ET+E+E + + ++++++ + + L L + + HS EY + K++
Sbjct 316 ANGIPPDTQKETSEQEQKTVKETLVSAYEKNISTKLKAELCNAESHSYEYTKAAAKNLKS 375
Query 74 -DILALLWRTFCFLDERGFKTQ 94
W F L +RG + +
Sbjct 376 EQWCTTWWYQFTVLLQRGVRER 397
> dre:100333349 nuclear receptor coactivator 2-like; K11256 nuclear
receptor coactivator 3 [EC:2.3.1.48]
Length=1497
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 34/118 (28%), Positives = 55/118 (46%), Gaps = 9/118 (7%)
Query 23 AITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRT 82
AI + T +I+QI +S DDD+ K+ S K+ LG +L
Sbjct 67 AILKETVRQIRQIKEQGKSSCG---DDDVQKADVSSTGQGVIDKDHLGP---LLLQALDG 120
Query 83 FCFLDERGFKTQAATVALKVNEHMHYKPDN-IAIKMWDVLRKELPEELCKDLPGENIP 139
F F+ R + V+ V +++ YK + I ++++L +E EEL K+LP N P
Sbjct 121 FLFVVSR--EGSIVFVSDNVTQYLQYKQEELINTSIYNILHEEDREELHKNLPKSNGP 176
> dre:564519 ncoa3, sb:eu248, sb:eu484, src3; nuclear receptor
coactivator 3; K11256 nuclear receptor coactivator 3 [EC:2.3.1.48]
Length=1375
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 34/118 (28%), Positives = 55/118 (46%), Gaps = 9/118 (7%)
Query 23 AITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSIDILALLWRT 82
AI + T +I+QI +S DDD+ K+ S K+ LG +L
Sbjct 67 AILKETVRQIRQIKEQGKSSCG---DDDVQKADVSSTGQGVIDKDHLGP---LLLQALDG 120
Query 83 FCFLDERGFKTQAATVALKVNEHMHYKPDN-IAIKMWDVLRKELPEELCKDLPGENIP 139
F F+ R + V+ V +++ YK + I ++++L +E EEL K+LP N P
Sbjct 121 FLFVVSR--EGSIVFVSDNVTQYLQYKQEELINTSIYNILHEEDREELHKNLPKSNGP 176
> sce:YOL115W PAP2, TRF4; Non-canonical poly(A) polymerase, involved
in nuclear RNA degradation as a component of the TRAMP
complex; catalyzes polyadenylation of hypomodified tRNAs,
and snoRNA and rRNA precursors; overlapping but non-redundant
functions with Trf5p (EC:2.7.7.-); K03514 DNA polymerase sigma
subunit [EC:2.7.7.7]
Length=584
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 30/57 (52%), Gaps = 5/57 (8%)
Query 59 DVHSDEYKERLGKSI--DILALLWRTFCFLDERGFKTQAATVALKVNEHMHYKPDNI 113
++HS +K+RLGKSI +++ + F DERG A + NE+ H K I
Sbjct 460 ELHSATFKDRLGKSILGNVIKYRGKARDFKDERGLVLNKAIIE---NENYHKKRSRI 513
> hsa:8416 ANXA9, ANX31; annexin A9
Length=345
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 11 LEATEAQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQS 57
L A QGV AI + ++ I +F ER DLMKSLQ+
Sbjct 51 LRAITGQGVDRSAIVDVLTNRSREQRQLISRNFQERTQQDLMKSLQA 97
> mmu:70575 Gfod2, 5730466C23Rik; glucose-fructose oxidoreductase
domain containing 2
Length=385
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 3/65 (4%)
Query 15 EAQGVTTEAITETTEEEIQQISASI-LTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSI 73
A+G T EA+ TEEE +Q++ + +T + R DD L+ Q +D+ L + I
Sbjct 24 RAEGFTVEALWGKTEEEAKQLAEEMNITFYTSRTDDVLLH--QDVDLVCINIPPPLTRQI 81
Query 74 DILAL 78
+ AL
Sbjct 82 SVKAL 86
> ath:AT1G10760 SEX1; SEX1 (STARCH EXCESS 1); alpha-glucan, water
dikinase; K08244 alpha-glucan, water dikinase [EC:2.7.9.4]
Length=1399
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 18 GVTTEAITETTEEEIQQISASILTSFVERLDDDLMKS 54
GV A++ TEE I+ SA+ L+S V RLD L K+
Sbjct 901 GVDQSAVSIFTEEIIRAGSAAALSSLVNRLDPVLRKT 937
> mmu:231769 Sfswap, 1190005N23Rik, 6330437E22Rik, AI197402, AW212079,
SWAP, Sfrs8, Srsf8; splicing factor, suppressor of
white-apricot homolog (Drosophila)
Length=945
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 8 AAGLEATEAQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQSI 58
+A + E++G + E ++E+ QIS++I++S ++ DLM ++++
Sbjct 883 SASISPVESRGSSQERSRGVSQEKDGQISSAIVSSVQSKITQDLMAKVRAM 933
> cel:T28A11.5 hypothetical protein
Length=194
Score = 28.5 bits (62), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 5/34 (14%)
Query 109 KPDNIAIKMWD-----VLRKELPEELCKDLPGEN 137
KP KMW +++ +LP+ELCK++ G++
Sbjct 124 KPSPECYKMWGPFDGFIIKDKLPDELCKNMVGKD 157
> dre:30544 sox17, MGC91776; SRY-box containing gene 17; K04495
transcription factor SOX17 (SOX group F)
Length=413
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query 56 QSIDVHSDEYKERLGKSIDILALLWRTFCFLDERGFKTQAATVALK-VNEHMHYK 109
Q+ D+H+ E + LGKS W+ +D+R F +A + +K + +H +YK
Sbjct 84 QNPDLHNAELSKMLGKS-------WKALPMVDKRPFVEEAERLRVKHMQDHPNYK 131
> hsa:81577 GFOD2, FLJ23802, MGC11335; glucose-fructose oxidoreductase
domain containing 2
Length=385
Score = 28.1 bits (61), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 1/64 (1%)
Query 15 EAQGVTTEAITETTEEEIQQISASILTSFVERLDDDLMKSLQSIDVHSDEYKERLGKSID 74
A+G T EA+ TEEE +Q++ + +F DD++ Q +D+ L + I
Sbjct 24 RAEGFTVEALWGKTEEEAKQLAEEMNIAFYTSRTDDILLH-QDVDLVCISIPPPLTRQIS 82
Query 75 ILAL 78
+ AL
Sbjct 83 VKAL 86
Lambda K H
0.315 0.132 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40