bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1446_orf1
Length=77
Score E
Sequences producing significant alignments: (Bits) Value
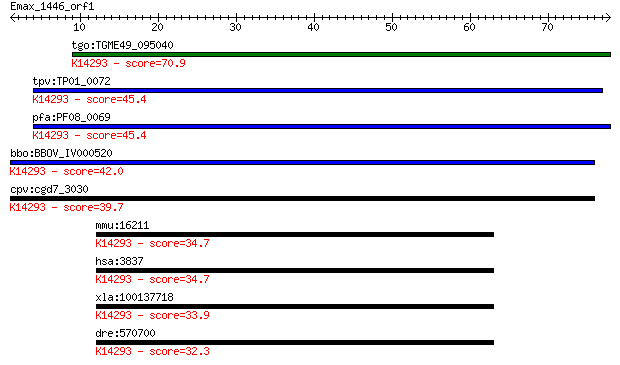
tgo:TGME49_095040 importin subunit beta-1, putative ; K14293 i... 70.9 9e-13
tpv:TP01_0072 importin beta; K14293 importin subunit beta-1 45.4 4e-05
pfa:PF08_0069 importin beta, putative; K14293 importin subunit... 45.4 5e-05
bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 imp... 42.0 4e-04
cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit b... 39.7 0.002
mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin (... 34.7 0.079
hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC... 34.7 0.080
xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopher... 33.9 0.12
dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopher... 32.3 0.34
> tgo:TGME49_095040 importin subunit beta-1, putative ; K14293
importin subunit beta-1
Length=971
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 48/69 (69%), Gaps = 9/69 (13%)
Query 9 SGELLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNPLDPIFP 68
S LL AIV RL DQPRVAV++CW+LHELAD++ +G + A+T PLDP+F
Sbjct 510 SNSLLAAIVRRLLDQPRVAVNVCWLLHELADHMTAGDGE---RPAST------PLDPLFQ 560
Query 69 KLCEALLSV 77
+LC+AL+ V
Sbjct 561 RLCDALIQV 569
> tpv:TP01_0072 importin beta; K14293 importin subunit beta-1
Length=873
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 40/73 (54%), Gaps = 11/73 (15%)
Query 4 GASNASGELLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNPL 63
G+ + L IV L D PRVAV+IC+ ++ELA++IN Y T N L
Sbjct 462 GSPDVPNSNLSKIVRALFDVPRVAVNICYFINELAEHIND------YNKGPT-----NLL 510
Query 64 DPIFPKLCEALLS 76
D +F +LCE L++
Sbjct 511 DCMFARLCEMLVN 523
> pfa:PF08_0069 importin beta, putative; K14293 importin subunit
beta-1
Length=877
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 7/74 (9%)
Query 4 GASNASGELLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNPL 63
G+ N S L ++ERL D PRVA ++CW+ ++LA N R +Y T + L
Sbjct 453 GSYNDSNSLYGILLERLNDYPRVAANVCWVFNQLAVN-----KRTSYNKMTNSYVTD--L 505
Query 64 DPIFPKLCEALLSV 77
D F LC+ L+ V
Sbjct 506 DDSFCVLCKKLIDV 519
> bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 importin
subunit beta-1
Length=859
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 12/75 (16%)
Query 1 PSGGASNASGELLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQ 60
P G+ G + I+ L RVA +ICW HELA+ + E +
Sbjct 443 PHLGSLEDPGSNICKIMRALFKPARVAANICWFFHELAEGLGQIEA------------SR 490
Query 61 NPLDPIFPKLCEALL 75
LD +FPK+C+AL+
Sbjct 491 YMLDAMFPKICDALV 505
> cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit
beta-1
Length=882
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 12/75 (16%)
Query 1 PSGGASNASGELLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQ 60
P +++G L L + LAD+PRV +ICW++H++A++ EG Q
Sbjct 460 PQQSVEHSNGLLSLLLQRLLADEPRVCTNICWIIHQIAESSQYVEG------------GQ 507
Query 61 NPLDPIFPKLCEALL 75
LD FP + ++LL
Sbjct 508 KVLDIWFPYIVQSLL 522
> mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 34.7 bits (78), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 8/51 (15%)
Query 12 LLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNP 62
LL ++E L+ +PRVA ++CW LA+ AAY+ A A + P
Sbjct 452 LLQCLIEGLSAEPRVASNVCWAFSSLAE--------AAYEAADVADDQEEP 494
> hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC2157,
NTF97; karyopherin (importin) beta 1; K14293 importin
subunit beta-1
Length=876
Score = 34.7 bits (78), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 8/51 (15%)
Query 12 LLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNP 62
LL ++E L+ +PRVA ++CW LA+ AAY+ A A + P
Sbjct 452 LLQCLIEGLSAEPRVASNVCWAFSSLAE--------AAYEAADVADDQEEP 494
> xla:100137718 kpnb1, imb1, impnb, ipo1, ipob, ntf97; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 8/51 (15%)
Query 12 LLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNP 62
LL ++E L +PRVA ++CW LA+ AAY+ A A + P
Sbjct 452 LLQCLIEGLGAEPRVASNVCWAFSSLAE--------AAYEAADVADDQEEP 494
> dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 8/51 (15%)
Query 12 LLLAIVERLADQPRVAVHICWMLHELADNINTPEGRAAYQGATTAQQDQNP 62
LL ++E L +PRVA ++CW LA+ AAY+ A + P
Sbjct 452 LLQCLIEGLGAEPRVASNVCWAFSSLAE--------AAYEATDVADDQEEP 494
Lambda K H
0.316 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072310352
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40