bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
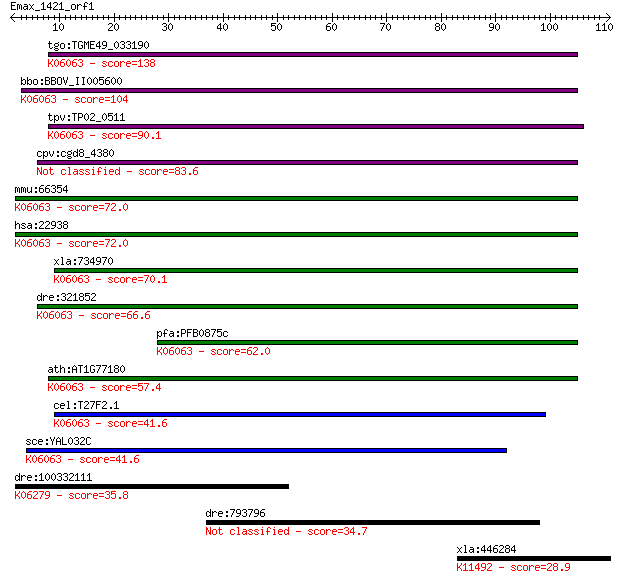
Query= Emax_1421_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033190 hypothetical protein ; K06063 SNW domain-con... 138 5e-33
bbo:BBOV_II005600 18.m06466; ski-interacting protein; K06063 S... 104 7e-23
tpv:TP02_0511 hypothetical protein; K06063 SNW domain-containi... 90.1 1e-18
cpv:cgd8_4380 SNW family nuclear protein 83.6 1e-16
mmu:66354 Snw1, 2310008B08Rik, AW048543, MGC55033, NCoA-62, SK... 72.0 4e-13
hsa:22938 SNW1, Bx42, MGC119379, NCOA-62, PRPF45, Prp45, SKIIP... 72.0 4e-13
xla:734970 snw1, MGC132028, bx42, ncoa-62, prp45, prpf45, skii... 70.1 2e-12
dre:321852 snw1, MGC123090, fb37b08, skiip, wu:fb37b08, zgc:12... 66.6 2e-11
pfa:PFB0875c chromatin-binding protein, putative; K06063 SNW d... 62.0 5e-10
ath:AT1G77180 chromatin protein family; K06063 SNW domain-cont... 57.4 1e-08
cel:T27F2.1 skp-1; mammalian SKIP (Ski interacting protein) ho... 41.6 6e-04
sce:YAL032C PRP45, FUN20; Protein required for pre-mRNA splici... 41.6 7e-04
dre:100332111 SHC (Src homology 2 domain containing) transform... 35.8 0.038
dre:793796 scg2a, fj60f08, wu:fj60f08; secretogranin II (chrom... 34.7 0.078
xla:446284 ncapg2, MGC130689; non-SMC condensin II complex, su... 28.9 4.3
> tgo:TGME49_033190 hypothetical protein ; K06063 SNW domain-containing
protein 1
Length=557
Score = 138 bits (347), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 65/98 (66%), Positives = 77/98 (78%), Gaps = 1/98 (1%)
Query 8 MFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQR-GGAGIYQFSRDRFASSVGENSEL 66
+FD RLFNQ G DSG+ GG D+ YNLYD+PLFA R A IYQFSR+R +SVG ++
Sbjct 459 VFDTRLFNQSAGVDSGFDGGNDEAYNLYDQPLFANRSNNAAIYQFSRERLINSVGHTGDV 518
Query 67 ASFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHK 104
SFAGAD++ +TRT PVEFEKDV+DPFGLDNLLSEA K
Sbjct 519 PSFAGADRSTFTRTAPVEFEKDVSDPFGLDNLLSEAKK 556
> bbo:BBOV_II005600 18.m06466; ski-interacting protein; K06063
SNW domain-containing protein 1
Length=458
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 53/105 (50%), Positives = 65/105 (61%), Gaps = 3/105 (2%)
Query 3 PQPDSMFDA---RLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQFSRDRFASS 59
P P + DA RL N G DSG+ GGED+ YN+YD+PLFA R GIYQ S +RF S
Sbjct 353 PVPSKLSDAHDTRLLNTAAGIDSGFDGGEDENYNIYDKPLFADRSIVGIYQHSSERFQQS 412
Query 60 VGENSELASFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHK 104
+G N + + A+ T RT PVEF K+ ADPFGL LL +A K
Sbjct 413 LGVNEAMRVPSFANATMVQRTTPVEFVKETADPFGLGTLLDKAKK 457
> tpv:TP02_0511 hypothetical protein; K06063 SNW domain-containing
protein 1
Length=455
Score = 90.1 bits (222), Expect = 1e-18, Method: Composition-based stats.
Identities = 47/98 (47%), Positives = 63/98 (64%), Gaps = 3/98 (3%)
Query 8 MFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQFSRDRFASSVGENSELA 67
++D RL G SG+ GG+D+ YN+YD+PLFA R A IYQ S++RF S G+ +A
Sbjct 361 LYDTRLLQGDAGISSGFDGGDDEGYNIYDKPLFADRSTANIYQHSKERFQKSTGD-VNVA 419
Query 68 SFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHKS 105
SFA AD+ R PVEF KD +DPFG + LL + K+
Sbjct 420 SFANADRG-VQRNTPVEFVKD-SDPFGFEKLLEKVKKT 455
> cpv:cgd8_4380 SNW family nuclear protein
Length=431
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/102 (44%), Positives = 60/102 (58%), Gaps = 6/102 (5%)
Query 6 DSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRG--GAGIYQFSRDRFASSVGEN 63
+S FDARLFN+ G DSG+ +DT ++YD+PLF G+Y F R S+G
Sbjct 329 ESQFDARLFNKVSGLDSGF---SNDTISVYDKPLFNTNNLKHRGLYTFEESRVEESIGGR 385
Query 64 SELASFAGADKTRYT-RTGPVEFEKDVADPFGLDNLLSEAHK 104
+ SF+G D ++ RT PVEFE+D DPFGLD L+ K
Sbjct 386 VHVPSFSGTDNSKTALRTKPVEFERDEDDPFGLDKLIDSVRK 427
> mmu:66354 Snw1, 2310008B08Rik, AW048543, MGC55033, NCoA-62,
SKIP, Skiip; SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/121 (38%), Positives = 66/121 (54%), Gaps = 24/121 (19%)
Query 2 IPQP----DSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGA----GIYQFSR 53
+P P + +D RLFNQ G DSG+ GGED+ YN+YD+ A RGG IY+ S+
Sbjct 396 VPNPRTSNEVQYDQRLFNQSKGMDSGFAGGEDEIYNVYDQ---AWRGGKDMAQSIYRPSK 452
Query 54 DRFASSVGENSEL----------ASFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAH 103
+ G++ E F+G+D+ + R GPV+FE+ DPFGLD L EA
Sbjct 453 NLDKDMYGDDLEARIKTNRFVPDKEFSGSDRKQRGREGPVQFEE---DPFGLDKFLEEAK 509
Query 104 K 104
+
Sbjct 510 Q 510
> hsa:22938 SNW1, Bx42, MGC119379, NCOA-62, PRPF45, Prp45, SKIIP,
SKIP; SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/121 (38%), Positives = 66/121 (54%), Gaps = 24/121 (19%)
Query 2 IPQP----DSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGA----GIYQFSR 53
+P P + +D RLFNQ G DSG+ GGED+ YN+YD+ A RGG IY+ S+
Sbjct 396 VPNPRTSNEVQYDQRLFNQSKGMDSGFAGGEDEIYNVYDQ---AWRGGKDMAQSIYRPSK 452
Query 54 DRFASSVGENSEL----------ASFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAH 103
+ G++ E F+G+D+ + R GPV+FE+ DPFGLD L EA
Sbjct 453 NLDKDMYGDDLEARIKTNRFVPDKEFSGSDRRQRGREGPVQFEE---DPFGLDKFLEEAK 509
Query 104 K 104
+
Sbjct 510 Q 510
> xla:734970 snw1, MGC132028, bx42, ncoa-62, prp45, prpf45, skiip;
SNW domain containing 1; K06063 SNW domain-containing protein
1
Length=535
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 45/107 (42%), Positives = 60/107 (56%), Gaps = 15/107 (14%)
Query 9 FDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGA-GIYQFSR--------DRFASS 59
+D RLFNQ G DSG+ GGED+ YN+YD+P + A IY+ S+ D +
Sbjct 407 YDQRLFNQSRGMDSGFAGGEDEVYNVYDQPWLGNKKLAQNIYRPSKNTDNDVYGDDLDTL 466
Query 60 VGENSEL--ASFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHK 104
V N + F+GAD+ R GPV+FE+ DPFGLD L EA +
Sbjct 467 VKTNRFVPDKDFSGADR-RQRHEGPVQFEE---DPFGLDKFLEEAKQ 509
> dre:321852 snw1, MGC123090, fb37b08, skiip, wu:fb37b08, zgc:123090;
SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 61/110 (55%), Gaps = 15/110 (13%)
Query 6 DSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGA-GIYQFSRDRFASSVGENS 64
++ +D RLFNQ G DSG+ GGED+ YN+YD+P R A IY+ S++ G++
Sbjct 406 EAQYDQRLFNQSKGMDSGFAGGEDEMYNVYDQPFRGGRDMAQNIYRPSKNVDKDMYGDDL 465
Query 65 ELA----------SFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHK 104
+ F+GAD R GPV+FE+ DPFGLD L EA +
Sbjct 466 DTLMQNNRFVPDRDFSGADHG-PRRDGPVQFEE---DPFGLDKFLEEAKQ 511
> pfa:PFB0875c chromatin-binding protein, putative; K06063 SNW
domain-containing protein 1
Length=482
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 33/77 (42%), Positives = 47/77 (61%), Gaps = 11/77 (14%)
Query 28 EDDTYNLYDRPLFAQRGGAGIYQFSRDRFASSVGENSELASFAGADKTRYTRTGPVEFEK 87
+DDTY +YD LF + A IY+FS +R +V + +TR T PV++ K
Sbjct 416 DDDTYQIYDTALFNNKNNANIYKFSSERLRKNVQK----------IETRDTMQ-PVKYIK 464
Query 88 DVADPFGLDNLLSEAHK 104
D++DPFGLD+LLS+A K
Sbjct 465 DISDPFGLDSLLSQAKK 481
> ath:AT1G77180 chromatin protein family; K06063 SNW domain-containing
protein 1
Length=613
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 44/113 (38%), Positives = 60/113 (53%), Gaps = 18/113 (15%)
Query 8 MFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLF-AQRGGAGIYQFSRDRFASSVG----- 61
M+D RLFNQ G DSG+ DD YNLYD+ LF AQ + +Y+ +D G
Sbjct 458 MYDQRLFNQDKGMDSGF--AADDQYNLYDKGLFTAQPTLSTLYKPKKDNDEEMYGNADEQ 515
Query 62 ----ENSEL----ASFAGA-DKTRYTRTGPVEFEK-DVADPFGLDNLLSEAHK 104
+N+E +F GA ++ R PVEFEK + DPFGL+ +S+ K
Sbjct 516 LDKIKNTERFKPDKAFTGASERVGSKRDRPVEFEKEEEQDPFGLEKWVSDLKK 568
> cel:T27F2.1 skp-1; mammalian SKIP (Ski interacting protein)
homolog family member (skp-1); K06063 SNW domain-containing
protein 1
Length=535
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 52/112 (46%), Gaps = 36/112 (32%)
Query 9 FDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQF----------------- 51
FD RLF++ G DSG +DDTYN YD A RGG + Q
Sbjct 412 FDQRLFDKTQGLDSG--AMDDDTYNPYD---AAWRGGDSVQQHVYRPSKNLDNDVYGGDL 466
Query 52 -----SRDRFASSVGENSELASFAGADKTRYTRTGPVEFEKDVADPFGLDNL 98
++RF + G F+GA+ + +GPV+FEKD D FGL +L
Sbjct 467 DKIIEQKNRFVADKG-------FSGAEGSSRG-SGPVQFEKD-QDVFGLSSL 509
> sce:YAL032C PRP45, FUN20; Protein required for pre-mRNA splicing;
associates with the spliceosome and interacts with splicing
factors Prp22p and Prp46p; orthologous to human transcriptional
coactivator SKIP and can activate transcription of
a reporter gene; K06063 SNW domain-containing protein 1
Length=379
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 44/88 (50%), Gaps = 14/88 (15%)
Query 4 QPDSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQFSRDRFASSVGEN 63
QPD +D+R F +G ++ K ED +YD PLF Q+ IY+ + ++ +V
Sbjct 298 QPDLQYDSRFFTRGA--NASAKRHEDQ---VYDNPLFVQQDIESIYKTNYEKLDEAVNVK 352
Query 64 SELASFAGADKTRYTRTGPVEFEKDVAD 91
SE AS + GP++F K +D
Sbjct 353 SEGASGSH---------GPIQFTKAESD 371
> dre:100332111 SHC (Src homology 2 domain containing) transforming
protein 3-like; K06279 src homology 2 domain-containing
transforming protein C
Length=393
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query 2 IPQPDSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQF 51
+P P DARL NQ DS G D TY RP+F Q+G IY
Sbjct 148 MPPPGGFIDARLTNQ--SQDSSQGAGVDQTY-YQGRPMFIQQGSCDIYSL 194
> dre:793796 scg2a, fj60f08, wu:fj60f08; secretogranin II (chromogranin
C) a
Length=539
Score = 34.7 bits (78), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 29/61 (47%), Gaps = 3/61 (4%)
Query 37 RPLFAQRGGAGIYQFSRDRFASSVGENSELASFAGADKTRYTRTGPVEFEKDVADPFGLD 96
RP A+ Q SRD + + GE ELA++ + R+TR P+ +D P L
Sbjct 433 RPTQARNERYYERQTSRDDYDDTAGEEDELANYLATEMQRHTRMAPL---RDEDQPVNLQ 489
Query 97 N 97
N
Sbjct 490 N 490
> xla:446284 ncapg2, MGC130689; non-SMC condensin II complex,
subunit G2; K11492 condensin-2 complex subunit G2
Length=1156
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 1/29 (3%)
Query 83 VEFEKDVADPFGLDNLLSE-AHKSPVEGW 110
++ KDV+DPF L+ LL E + K E W
Sbjct 23 IQLHKDVSDPFDLNELLQELSRKQKEELW 51
Lambda K H
0.314 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40