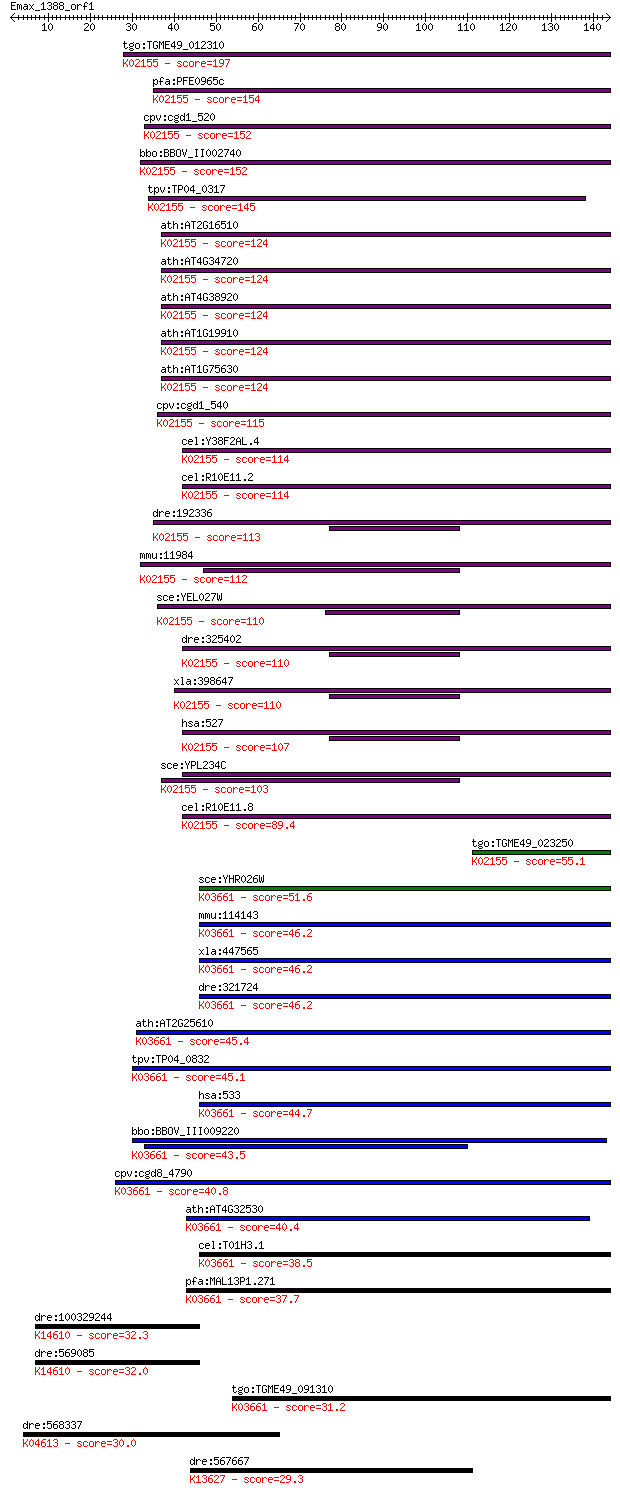
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1388_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_012310 vacuolar ATP synthase 16 kDa proteolipid sub... 197 1e-50
pfa:PFE0965c vacuolar ATP synthetase; K02155 V-type H+-transpo... 154 7e-38
cpv:cgd1_520 vacuolar ATP synthetase subunit ; K02155 V-type H... 152 5e-37
bbo:BBOV_II002740 18.m06223; proteolipid subunit c; K02155 V-t... 152 5e-37
tpv:TP04_0317 vacuolar ATPase subunit (EC:3.6.3.14); K02155 V-... 145 6e-35
ath:AT2G16510 vacuolar ATP synthase 16 kDa proteolipid subunit... 124 1e-28
ath:AT4G34720 AVA-P1; AVA-P1; ATPase/ proton-transporting ATPa... 124 1e-28
ath:AT4G38920 ATVHA-C3 (VACUOLAR-TYPE H(+)-ATPASE C3); ATPase;... 124 1e-28
ath:AT1G19910 AVA-P2; AVA-P2; ATPase/ proton-transporting ATPa... 124 1e-28
ath:AT1G75630 AVA-P4; AVA-P4; ATPase; K02155 V-type H+-transpo... 124 1e-28
cpv:cgd1_540 vacuolar ATP synthase subunit, possible signal pe... 115 5e-26
cel:Y38F2AL.4 vha-3; Vacuolar H ATPase family member (vha-3); ... 114 1e-25
cel:R10E11.2 vha-2; Vacuolar H ATPase family member (vha-2); K... 114 1e-25
dre:192336 atp6v0ca, CHUNP6904, MGC174785, atp6l, atp6v0c, cb9... 113 2e-25
mmu:11984 Atp6v0c, Atp6c, Atp6c2, Atp6l, Atpl, Atpl-rs1, PL16,... 112 4e-25
sce:YEL027W CUP5, CLS7, GEF2, VMA3; Cup5p (EC:3.6.3.14); K0215... 110 1e-24
dre:325402 atp6v0cb, MGC111904, wu:fc74d11, zgc:111904, zgc:77... 110 1e-24
xla:398647 atp6v0c, MGC64475, atp6c, atp6l, atpl, vatl, vma3; ... 110 2e-24
hsa:527 ATP6V0C, ATP6C, ATP6L, ATPL, VATL, VPPC, Vma3; ATPase,... 107 1e-23
sce:YPL234C TFP3, CLS9, VMA11; Tfp3p (EC:3.6.3.14); K02155 V-t... 103 3e-22
cel:R10E11.8 vha-1; Vacuolar H ATPase family member (vha-1); K... 89.4 4e-18
tgo:TGME49_023250 vacuolar type H+-ATPase proteolipid subunit,... 55.1 7e-08
sce:YHR026W PPA1, VMA16; Ppa1p (EC:3.6.3.14); K03661 V-type H+... 51.6 8e-07
mmu:114143 Atp6v0b, 2310024H13Rik, Atp6f, VMA16; ATPase, H+ tr... 46.2 3e-05
xla:447565 atp6v0b, MGC84266, atp6f, hatpl, vma16; ATPase, H+ ... 46.2 4e-05
dre:321724 atp6v0b, wu:fb33a01, zgc:63832; ATPase, H+ transpor... 46.2 4e-05
ath:AT2G25610 H+-transporting two-sector ATPase, C subunit fam... 45.4 6e-05
tpv:TP04_0832 vacuolar ATP synthase subunit C (EC:3.6.3.14); K... 45.1 8e-05
hsa:533 ATP6V0B, ATP6F, HATPL, VMA16; ATPase, H+ transporting,... 44.7 9e-05
bbo:BBOV_III009220 17.m10611; ATP synthase subunit C domain co... 43.5 2e-04
cpv:cgd8_4790 proteolipid subunit of the vacuolar ATpase ; K03... 40.8 0.001
ath:AT4G32530 vacuolar ATP synthase, putative / V-ATPase, puta... 40.4 0.002
cel:T01H3.1 vha-4; Vacuolar H ATPase family member (vha-4); K0... 38.5 0.007
pfa:MAL13P1.271 V-type ATPase, putative; K03661 V-type H+-tran... 37.7 0.013
dre:100329244 solute carrier family 19 (thiamine transporter),... 32.3 0.53
dre:569085 solute carrier family 19 (thiamine transporter), me... 32.0 0.63
tgo:TGME49_091310 vacuolar ATP synthase 22 kDa proteolipid sub... 31.2 1.2
dre:568337 olfcd3, VR3.13a, v2r2l2; olfactory receptor C famil... 30.0 2.8
dre:567667 slc12a7b, sb:cb734; solute carrier family 12, membe... 29.3 4.1
> tgo:TGME49_012310 vacuolar ATP synthase 16 kDa proteolipid subunit,
putative (EC:3.6.3.14); K02155 V-type H+-transporting
ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=170
Score = 197 bits (500), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 96/116 (82%), Positives = 109/116 (93%), Gaps = 0/116 (0%)
Query 28 MFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILP 87
MFWSAFLQCDPNS FFGFMGIT+A+VF+NLGAAYGTAKSGVG+SS+GVMRPDL+MRSI+P
Sbjct 1 MFWSAFLQCDPNSTFFGFMGITAAMVFSNLGAAYGTAKSGVGISSMGVMRPDLVMRSIIP 60
Query 88 VIMAGILGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
V+MAGILGIYGLIISIVI+ M +PD YS+YAG+GHLA GL VGLSA+AAGLAIGI
Sbjct 61 VVMAGILGIYGLIISIVINGSMDTPDTYSSYAGYGHLAAGLTVGLSAMAAGLAIGI 116
> pfa:PFE0965c vacuolar ATP synthetase; K02155 V-type H+-transporting
ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=165
Score = 154 bits (390), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 78/109 (71%), Positives = 94/109 (86%), Gaps = 0/109 (0%)
Query 35 QCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGIL 94
QCDPNS FFGFMGI ++ +F+NLGAA+GTAKSGVGV SVGVMRPDLIM+SILPV+MAG+L
Sbjct 3 QCDPNSAFFGFMGIAASSIFSNLGAAFGTAKSGVGVCSVGVMRPDLIMKSILPVVMAGVL 62
Query 95 GIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
GIYG+I+SI+I M + YS +AG+ HL+ GL VGLS+LAAGLAIGI
Sbjct 63 GIYGIIMSILIYGKMTPAEGYSTFAGYAHLSSGLIVGLSSLAAGLAIGI 111
> cpv:cgd1_520 vacuolar ATP synthetase subunit ; K02155 V-type
H+-transporting ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=165
Score = 152 bits (383), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 75/111 (67%), Positives = 92/111 (82%), Gaps = 0/111 (0%)
Query 33 FLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAG 92
+ CDPNS+FFGF+GI ++FANLGAAYG AKSGVG+SS+ VMRPDLIMRSI+P +MAG
Sbjct 1 MITCDPNSLFFGFLGIAGCLIFANLGAAYGIAKSGVGISSMAVMRPDLIMRSIIPAVMAG 60
Query 93 ILGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
ILGIYGLI S+VI MG P++YSAY + ++ GL +GLS+LAAGLAIGI
Sbjct 61 ILGIYGLIGSLVIFFQMGEPNLYSAYTAYAQMSAGLVIGLSSLAAGLAIGI 111
> bbo:BBOV_II002740 18.m06223; proteolipid subunit c; K02155 V-type
H+-transporting ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=173
Score = 152 bits (383), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 76/112 (67%), Positives = 96/112 (85%), Gaps = 0/112 (0%)
Query 32 AFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMA 91
A + CDP+S+FFG MG S++VF++LGAAYGTA+SGVG+SS+GVMRPDL+MRSI+PVIMA
Sbjct 8 AKIPCDPHSIFFGLMGAVSSMVFSSLGAAYGTARSGVGISSMGVMRPDLVMRSIIPVIMA 67
Query 92 GILGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
G+LGIYGLI++++I MG P YSAYAG+ HL+ GL VG S LA+GLAIGI
Sbjct 68 GVLGIYGLIMAVIIVLNMGHPGSYSAYAGYSHLSAGLIVGFSGLASGLAIGI 119
> tpv:TP04_0317 vacuolar ATPase subunit (EC:3.6.3.14); K02155
V-type H+-transporting ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=119
Score = 145 bits (365), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 69/104 (66%), Positives = 88/104 (84%), Gaps = 0/104 (0%)
Query 34 LQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGI 93
++CDP+S+FFG MG+ A+VF+NLGAAYGTA+SGVG+SS+GVMRPDL+M+SI+PVIMAG+
Sbjct 3 VECDPHSIFFGMMGVVCAMVFSNLGAAYGTARSGVGISSMGVMRPDLVMKSIIPVIMAGV 62
Query 94 LGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAA 137
LGIYGLIISIVI+ G P YS + G+ HLA GL VGL +L +
Sbjct 63 LGIYGLIISIVITGNYGEPGEYSHFLGYSHLAAGLVVGLCSLVS 106
> ath:AT2G16510 vacuolar ATP synthase 16 kDa proteolipid subunit
5 / V-ATPase 16 kDa proteolipid subunit 5 (AVAP5); K02155
V-type H+-transporting ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=164
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 90/109 (82%), Gaps = 3/109 (2%)
Query 37 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 96
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 7 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 66
Query 97 YGLIISIVISSVMGSPDVYSAY--AGFGHLAGGLAVGLSALAAGLAIGI 143
YGLII+++IS+ + +P S Y G+ HL+ GLA GL+ L+AG+AIGI
Sbjct 67 YGLIIAVIISTGI-NPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGI 114
> ath:AT4G34720 AVA-P1; AVA-P1; ATPase/ proton-transporting ATPase,
rotational mechanism; K02155 V-type H+-transporting ATPase
16kDa proteolipid subunit [EC:3.6.3.14]
Length=164
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 90/109 (82%), Gaps = 3/109 (2%)
Query 37 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 96
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 7 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 66
Query 97 YGLIISIVISSVMGSPDVYSAY--AGFGHLAGGLAVGLSALAAGLAIGI 143
YGLII+++IS+ + +P S Y G+ HL+ GLA GL+ L+AG+AIGI
Sbjct 67 YGLIIAVIISTGI-NPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGI 114
> ath:AT4G38920 ATVHA-C3 (VACUOLAR-TYPE H(+)-ATPASE C3); ATPase;
K02155 V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=164
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 90/109 (82%), Gaps = 3/109 (2%)
Query 37 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 96
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 7 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 66
Query 97 YGLIISIVISSVMGSPDVYSAY--AGFGHLAGGLAVGLSALAAGLAIGI 143
YGLII+++IS+ + +P S Y G+ HL+ GLA GL+ L+AG+AIGI
Sbjct 67 YGLIIAVIISTGI-NPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGI 114
> ath:AT1G19910 AVA-P2; AVA-P2; ATPase/ proton-transporting ATPase,
rotational mechanism; K02155 V-type H+-transporting ATPase
16kDa proteolipid subunit [EC:3.6.3.14]
Length=165
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 90/109 (82%), Gaps = 3/109 (2%)
Query 37 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 96
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 8 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 67
Query 97 YGLIISIVISSVMGSPDVYSAY--AGFGHLAGGLAVGLSALAAGLAIGI 143
YGLII+++IS+ + +P S Y G+ HL+ GLA GL+ L+AG+AIGI
Sbjct 68 YGLIIAVIISTGI-NPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGI 115
> ath:AT1G75630 AVA-P4; AVA-P4; ATPase; K02155 V-type H+-transporting
ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=166
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 90/109 (82%), Gaps = 3/109 (2%)
Query 37 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 96
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 9 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 68
Query 97 YGLIISIVISSVMGSPDVYSAY--AGFGHLAGGLAVGLSALAAGLAIGI 143
YGLII+++IS+ + +P S Y G+ HL+ GLA GL+ L+AG+AIGI
Sbjct 69 YGLIIAVIISTGI-NPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGI 116
> cpv:cgd1_540 vacuolar ATP synthase subunit, possible signal
peptide ; K02155 V-type H+-transporting ATPase 16kDa proteolipid
subunit [EC:3.6.3.14]
Length=167
Score = 115 bits (288), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 57/108 (52%), Positives = 76/108 (70%), Gaps = 2/108 (1%)
Query 36 CDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILG 95
C P + FG +G T A +N GAAYGTAK+G+ ++S GVMRPDL+MRSI+P +MAGILG
Sbjct 8 CTPTATLFGMLGSTLATALSNFGAAYGTAKAGLAIASCGVMRPDLVMRSIIPAVMAGILG 67
Query 96 IYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+YGLI+ ++I S + + YS Y G+ HLA GL G S A+G IG+
Sbjct 68 VYGLIVGVIICSQIRTD--YSLYQGYCHLAAGLISGFSCAASGFTIGV 113
> cel:Y38F2AL.4 vha-3; Vacuolar H ATPase family member (vha-3);
K02155 V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=161
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 76/103 (73%), Gaps = 1/103 (0%)
Query 42 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 101
FFG+MG SA +F LGAAYGTAKS VG+ S+GVMRP+LIM+S++PVIMAGI+GIYGL++
Sbjct 16 FFGYMGAASAQIFTVLGAAYGTAKSAVGICSMGVMRPELIMKSVIPVIMAGIIGIYGLVV 75
Query 102 SIVISSVMGSPDV-YSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
++V+ + S Y GF HLA GL GL L AG AIGI
Sbjct 76 AMVLKGKVTSASAGYDLNKGFAHLAAGLTCGLCGLGAGYAIGI 118
> cel:R10E11.2 vha-2; Vacuolar H ATPase family member (vha-2);
K02155 V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=161
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 76/103 (73%), Gaps = 1/103 (0%)
Query 42 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 101
FFG+MG SA +F LGAAYGTAKS VG+ S+GVMRP+LIM+S++PVIMAGI+GIYGL++
Sbjct 16 FFGYMGAASAQIFTVLGAAYGTAKSAVGICSMGVMRPELIMKSVIPVIMAGIIGIYGLVV 75
Query 102 SIVISSVMGSPDV-YSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
++V+ + S Y GF HLA GL GL L AG AIGI
Sbjct 76 AMVLKGKVTSASAGYDLNKGFAHLAAGLTCGLCGLGAGYAIGI 118
> dre:192336 atp6v0ca, CHUNP6904, MGC174785, atp6l, atp6v0c, cb993,
fb57d09, wu:fb57d09; ATPase, H+ transporting, lysosomal,
V0 subunit c, a (EC:3.6.3.14); K02155 V-type H+-transporting
ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=154
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 62/109 (56%), Positives = 83/109 (76%), Gaps = 2/109 (1%)
Query 35 QCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGIL 94
Q S FF MG +SA+VF+ LGAAYGTAKSG ++++ VMRP+LIM+SI+PV+MAGI+
Sbjct 4 QNPQYSPFFAVMGASSAMVFSALGAAYGTAKSGTDIAAMSVMRPELIMKSIIPVVMAGII 63
Query 95 GIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
IYGL+++++I++ +G D S Y F HL GL+VGLS LAAG AIGI
Sbjct 64 AIYGLVVAVLIANNIG--DKISLYKSFLHLGAGLSVGLSGLAAGFAIGI 110
Score = 28.9 bits (63), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 77 RPDLIMRSILPVIMAGILGIYGLIISIVISS 107
+P L + IL +I A +LG+YGLI+++++S+
Sbjct 122 QPRLFVGMILILIFAEVLGLYGLIVALILST 152
> mmu:11984 Atp6v0c, Atp6c, Atp6c2, Atp6l, Atpl, Atpl-rs1, PL16,
VATL, Vma3; ATPase, H+ transporting, lysosomal V0 subunit
C (EC:3.6.3.14); K02155 V-type H+-transporting ATPase 16kDa
proteolipid subunit [EC:3.6.3.14]
Length=155
Score = 112 bits (280), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 61/113 (53%), Positives = 87/113 (76%), Gaps = 3/113 (2%)
Query 32 AFLQCDPN-SVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIM 90
A ++ +P S FFG MG +SA+VF+ +GAAYGTAKSG G++++ VMRP+LIM+SI+PV+M
Sbjct 2 ADIKNNPEYSSFFGVMGASSAMVFSAMGAAYGTAKSGTGIAAMSVMRPELIMKSIIPVVM 61
Query 91 AGILGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
AGI+ IYGL+++++I++ + D + Y F L GL+VGLS LAAG AIGI
Sbjct 62 AGIIAIYGLVVAVLIANSL--TDGITLYRSFLQLGAGLSVGLSGLAAGFAIGI 112
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 37/72 (51%), Gaps = 11/72 (15%)
Query 47 GITSAIVFANLGAAYGTAKSGVG-------VSSVGV----MRPDLIMRSILPVIMAGILG 95
GIT F LGA SG+ V GV +P L + IL +I A +LG
Sbjct 83 GITLYRSFLQLGAGLSVGLSGLAAGFAIGIVGDAGVRGTAQQPRLFVGMILILIFAEVLG 142
Query 96 IYGLIISIVISS 107
+YGLI+++++S+
Sbjct 143 LYGLIVALILST 154
> sce:YEL027W CUP5, CLS7, GEF2, VMA3; Cup5p (EC:3.6.3.14); K02155
V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=160
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/108 (54%), Positives = 79/108 (73%), Gaps = 2/108 (1%)
Query 36 CDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILG 95
C + FFG +G SAI+F +LGAAYGTAKSGVG+ + V+RPDL+ ++I+PVIMAGI+
Sbjct 5 CPVYAPFFGAIGCASAIIFTSLGAAYGTAKSGVGICATCVLRPDLLFKNIVPVIMAGIIA 64
Query 96 IYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
IYGL++S+++ +G + Y GF L GL+VGLS LAAG AIGI
Sbjct 65 IYGLVVSVLVCYSLGQKQ--ALYTGFIQLGAGLSVGLSGLAAGFAIGI 110
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 76 MRPDLIMRSILPVIMAGILGIYGLIISIVISS 107
+P L + IL +I A +LG+YGLI++++++S
Sbjct 121 QQPRLFVGMILILIFAEVLGLYGLIVALLLNS 152
> dre:325402 atp6v0cb, MGC111904, wu:fc74d11, zgc:111904, zgc:77708;
ATPase, H+ transporting, lysosomal, V0 subunit c, b;
K02155 V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=153
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 58/102 (56%), Positives = 81/102 (79%), Gaps = 2/102 (1%)
Query 42 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 101
FF MG ++A+VF+ LGAAYGTAKSG G++++ VMRP+LIM+SI+PV+MAGI+ IYGL++
Sbjct 11 FFAVMGASAAMVFSALGAAYGTAKSGTGIAAMSVMRPELIMKSIIPVVMAGIIAIYGLVV 70
Query 102 SIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+++I++ + D + Y F HL GL+VGLS LAAG AIGI
Sbjct 71 AVLIANSI--SDKITLYKSFLHLGAGLSVGLSGLAAGFAIGI 110
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 77 RPDLIMRSILPVIMAGILGIYGLIISIVISS 107
+P L + IL +I A +LG+YGLI+++++S+
Sbjct 122 QPRLFVGMILILIFAEVLGLYGLIVALILST 152
> xla:398647 atp6v0c, MGC64475, atp6c, atp6l, atpl, vatl, vma3;
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c; K02155
V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=156
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/104 (55%), Positives = 80/104 (76%), Gaps = 2/104 (1%)
Query 40 SVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGL 99
S FF MG +SA+VF+ LGAAYGTAKSG G++++ VMRP+LIM+SI+PV+MAGI+ IYGL
Sbjct 12 SAFFAVMGASSAMVFSALGAAYGTAKSGTGIAAMSVMRPELIMKSIIPVVMAGIIAIYGL 71
Query 100 IISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+++++I++ + + Y F L GL+VGLS LAAG AIGI
Sbjct 72 VVAVLIANSL--TQTITLYKSFLQLGAGLSVGLSGLAAGFAIGI 113
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 77 RPDLIMRSILPVIMAGILGIYGLIISIVISS 107
+P L + IL +I A +LG+YGLI+++++S+
Sbjct 125 QPRLFVGMILILIFAEVLGLYGLIVALILST 155
> hsa:527 ATP6V0C, ATP6C, ATP6L, ATPL, VATL, VPPC, Vma3; ATPase,
H+ transporting, lysosomal 16kDa, V0 subunit c (EC:3.6.3.14);
K02155 V-type H+-transporting ATPase 16kDa proteolipid
subunit [EC:3.6.3.14]
Length=155
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/102 (55%), Positives = 79/102 (77%), Gaps = 2/102 (1%)
Query 42 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 101
FF MG ++A+VF+ LGAAYGTAKSG G++++ VMRP+ IM+SI+PV+MAGI+ IYGL++
Sbjct 13 FFAVMGASAAMVFSALGAAYGTAKSGTGIAAMSVMRPEQIMKSIIPVVMAGIIAIYGLVV 72
Query 102 SIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+++I++ + D S Y F L GL+VGLS LAAG AIGI
Sbjct 73 AVLIANSLN--DDISLYKSFLQLGAGLSVGLSGLAAGFAIGI 112
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 77 RPDLIMRSILPVIMAGILGIYGLIISIVISS 107
+P L + IL +I A +LG+YGLI+++++S+
Sbjct 124 QPRLFVGMILILIFAEVLGLYGLIVALILST 154
> sce:YPL234C TFP3, CLS9, VMA11; Tfp3p (EC:3.6.3.14); K02155 V-type
H+-transporting ATPase 16kDa proteolipid subunit [EC:3.6.3.14]
Length=164
Score = 103 bits (256), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 49/102 (48%), Positives = 78/102 (76%), Gaps = 0/102 (0%)
Query 42 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 101
FFGF G +A+V + LGAA GTAKSG+G++ +G +P+LIM+S++PV+M+GIL IYGL++
Sbjct 17 FFGFAGCAAAMVLSCLGAAIGTAKSGIGIAGIGTFKPELIMKSLIPVVMSGILAIYGLVV 76
Query 102 SIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+++I+ + + Y+ + GF HL+ GL VG + L++G AIG+
Sbjct 77 AVLIAGNLSPTEDYTLFNGFMHLSCGLCVGFACLSSGYAIGM 118
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 44/74 (59%), Gaps = 4/74 (5%)
Query 37 DPNSVFFGFMGITSAIV--FANLGAAYGTAKSG-VGVSSVGVMRPDLIMRSILPVIMAGI 93
+ ++F GFM ++ + FA L + Y G VGV + +P L + +L +I + +
Sbjct 88 EDYTLFNGFMHLSCGLCVGFACLSSGYAIGMVGDVGVRKY-MHQPRLFVGIVLILIFSEV 146
Query 94 LGIYGLIISIVISS 107
LG+YG+I+++++++
Sbjct 147 LGLYGMIVALILNT 160
> cel:R10E11.8 vha-1; Vacuolar H ATPase family member (vha-1);
K02155 V-type H+-transporting ATPase 16kDa proteolipid subunit
[EC:3.6.3.14]
Length=169
Score = 89.4 bits (220), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 53/103 (51%), Positives = 75/103 (72%), Gaps = 1/103 (0%)
Query 42 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 101
FFG +G+TSA+ FA G+AYGTAK+G G++S+ V RPDL+M++I+PV+MAGI+ IYGL++
Sbjct 24 FFGSLGVTSAMAFAAAGSAYGTAKAGTGIASMAVARPDLVMKAIIPVVMAGIVAIYGLVV 83
Query 102 SIVIS-SVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
++++S V + Y+ F AGGL GL L AG AIGI
Sbjct 84 AVIVSGKVEPAGANYTINNAFSQFAGGLVCGLCGLGAGYAIGI 126
> tgo:TGME49_023250 vacuolar type H+-ATPase proteolipid subunit,
putative (EC:3.6.3.14); K02155 V-type H+-transporting ATPase
16kDa proteolipid subunit [EC:3.6.3.14]
Length=165
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 26/33 (78%), Positives = 30/33 (90%), Gaps = 0/33 (0%)
Query 111 SPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+PD YS+YAG+GHLA GL VGLSA+AAGLAIGI
Sbjct 79 TPDTYSSYAGYGHLAAGLTVGLSAMAAGLAIGI 111
> sce:YHR026W PPA1, VMA16; Ppa1p (EC:3.6.3.14); K03661 V-type
H+-transporting ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=213
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 58/104 (55%), Gaps = 6/104 (5%)
Query 46 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 105
+GI + + +GAA+G +G + GV P + ++++ +I ++ IYGLII+IV
Sbjct 62 LGIALCVGLSVVGAAWGIFITGSSMIGAGVRAPRITTKNLISIIFCEVVAIYGLIIAIVF 121
Query 106 SS---VMGSPDVYSA---YAGFGHLAGGLAVGLSALAAGLAIGI 143
SS V + ++YS Y G+ G+ VG S L G+A+GI
Sbjct 122 SSKLTVATAENMYSKSNLYTGYSLFWAGITVGASNLICGIAVGI 165
> mmu:114143 Atp6v0b, 2310024H13Rik, Atp6f, VMA16; ATPase, H+
transporting, lysosomal V0 subunit B (EC:3.6.3.14); K03661 V-type
H+-transporting ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=205
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 59/109 (54%), Gaps = 14/109 (12%)
Query 46 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 105
+GI AI + +GAA+G +G + GV P + ++++ +I + IYG+I++IVI
Sbjct 52 LGIGLAISLSVVGAAWGIYITGSSIIGGGVKAPRIKTKNLVSIIFCEAVAIYGIIMAIVI 111
Query 106 SSVMGSPDVYSA-----------YAGFGHLAGGLAVGLSALAAGLAIGI 143
S+ M P +SA +AG+ GL VGLS L G+ +GI
Sbjct 112 SN-MAEP--FSATEPKAIGHRNYHAGYSMFGAGLTVGLSNLFCGVCVGI 157
> xla:447565 atp6v0b, MGC84266, atp6f, hatpl, vma16; ATPase, H+
transporting, lysosomal 21kDa, V0 subunit b (EC:3.6.3.14);
K03661 V-type H+-transporting ATPase 21kDa proteolipid subunit
[EC:3.6.3.14]
Length=205
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 58/106 (54%), Gaps = 8/106 (7%)
Query 46 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 105
+GI AI + +GAA+G +G + GV P + ++++ +I + IYG+I++IVI
Sbjct 52 LGIGLAISLSVVGAAWGIYITGSSILGGGVKAPRIKTKNLVSIIFCEAVAIYGIIMAIVI 111
Query 106 SSVMG-----SPDV---YSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
S++ +P+ + +AGF GL VG S L G+ +GI
Sbjct 112 SNMAEQFKGTTPEAIGNRNYHAGFSMFGAGLTVGFSNLFCGICVGI 157
> dre:321724 atp6v0b, wu:fb33a01, zgc:63832; ATPase, H+ transporting,
V0 subunit B (EC:3.6.3.14); K03661 V-type H+-transporting
ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=205
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 59/109 (54%), Gaps = 14/109 (12%)
Query 46 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 105
+GI AI + +GAA+G +G + GV P + ++++ +I + IYG+I++IVI
Sbjct 52 LGIGLAISLSVVGAAWGIYITGSSIIGGGVKAPRIKTKNLVSIIFCEAVAIYGIIMAIVI 111
Query 106 SSV-----------MGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
S++ +GS + + Y+ FG GL VG S L G+ +GI
Sbjct 112 SNLAENFSGTTPETIGSKNYQAGYSMFG---AGLTVGFSNLFCGICVGI 157
> ath:AT2G25610 H+-transporting two-sector ATPase, C subunit family
protein; K03661 V-type H+-transporting ATPase 21kDa proteolipid
subunit [EC:3.6.3.14]
Length=178
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 62/119 (52%), Gaps = 8/119 (6%)
Query 31 SAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIM 90
+A ++ P + F +GI +I + LGAA+G +G + + P + ++++ VI
Sbjct 13 AALVRISPYT--FSAIGIAISIGVSVLGAAWGIYITGSSLIGAAIEAPRITSKNLISVIF 70
Query 91 AGILGIYGLIISIVISSVMGSP------DVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+ IYG+I++I++ + + S D S AG+ A G+ VG + L GL +GI
Sbjct 71 CEAVAIYGVIVAIILQTKLESVPSSKMYDAESLRAGYAIFASGIIVGFANLVCGLCVGI 129
> tpv:TP04_0832 vacuolar ATP synthase subunit C (EC:3.6.3.14);
K03661 V-type H+-transporting ATPase 21kDa proteolipid subunit
[EC:3.6.3.14]
Length=179
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 35/131 (26%), Positives = 65/131 (49%), Gaps = 18/131 (13%)
Query 30 WSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVI 89
WS L+ D + F+G++GI ++ + GAA G G + V P + +++++ VI
Sbjct 5 WSTILK-DLSPSFWGYLGIFFSLGLSVFGAATGLMLCGPSIMGGSVKSPRITVKNLVSVI 63
Query 90 MAGILGIYGLIISIVISSV------------MGSPDVYSAY-----AGFGHLAGGLAVGL 132
+GIYGLI+S+++ ++ + ++ Y G+ A GL VG
Sbjct 64 FCEAIGIYGLIVSVLLMNIASRFTGEKAPLNLLDKEITKLYYNDLFRGYSMFAVGLIVGF 123
Query 133 SALAAGLAIGI 143
S L G+++G+
Sbjct 124 SNLFCGISVGV 134
> hsa:533 ATP6V0B, ATP6F, HATPL, VMA16; ATPase, H+ transporting,
lysosomal 21kDa, V0 subunit b (EC:3.6.3.14); K03661 V-type
H+-transporting ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=158
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 59/109 (54%), Gaps = 14/109 (12%)
Query 46 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 105
+GI AI + +GAA+G +G + GV P + ++++ +I + IYG+I++IVI
Sbjct 5 LGIGLAISLSVVGAAWGIYITGSSIIGGGVKAPRIKTKNLVSIIFCEAVAIYGIIMAIVI 64
Query 106 SSVMGSPDVYSA-----------YAGFGHLAGGLAVGLSALAAGLAIGI 143
S+ M P +SA +AG+ GL VGLS L G+ +GI
Sbjct 65 SN-MAEP--FSATDPKAIGHRNYHAGYSMFGAGLTVGLSNLFCGVCVGI 110
> bbo:BBOV_III009220 17.m10611; ATP synthase subunit C domain
containing protein; K03661 V-type H+-transporting ATPase 21kDa
proteolipid subunit [EC:3.6.3.14]
Length=180
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/131 (29%), Positives = 66/131 (50%), Gaps = 19/131 (14%)
Query 30 WSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVI 89
WS+ P S F+G++G A+ A LGA +G G + V P + +++++ VI
Sbjct 5 WSSVFANIPPS-FWGYLGTFLALGLAVLGAGWGILLCGTSIMGGSVNSPRITVKNLVSVI 63
Query 90 MAGILGIYGLIISI-VISSVMG-----SPDVYSA------------YAGFGHLAGGLAVG 131
+GIYGLI+++ ++++ +G PD ++A + G+ A GL G
Sbjct 64 FCEAVGIYGLIVAVLLLNASLGFTATPRPDDFNADKRITLLYFLEVHRGYVLFAIGLTSG 123
Query 132 LSALAAGLAIG 142
L L GL++G
Sbjct 124 LCNLFCGLSVG 134
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 41/77 (53%), Gaps = 3/77 (3%)
Query 33 FLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAG 92
FL+ V F +G+TS + G + G S ++ +P L ++ ++ I AG
Sbjct 106 FLEVHRGYVLFA-IGLTSGLCNLFCGLSVGAVGSACALADA--QKPQLFVKILMVEIFAG 162
Query 93 ILGIYGLIISIVISSVM 109
I+G++G+I ++++ S M
Sbjct 163 IIGLFGVIFAVLLLSTM 179
> cpv:cgd8_4790 proteolipid subunit of the vacuolar ATpase ; K03661
V-type H+-transporting ATPase 21kDa proteolipid subunit
[EC:3.6.3.14]
Length=181
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 62/132 (46%), Gaps = 16/132 (12%)
Query 26 FKMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSI 85
++ + FL P + F ++G+ IV + GA +G +G + + P + +++
Sbjct 3 YQTYSEIFLSIPP--LHFAYLGVVLCIVLSTFGAGWGIFTTGNSLVGAALRSPRIRSKNL 60
Query 86 LPVIMAGILGIYGLIIS-IVISSVMGSPD-------------VYSAYAGFGHLAGGLAVG 131
+ VI IYG+I + +++S + PD V + + L GL +G
Sbjct 61 ISVIFCEATAIYGVIATFLLMSKIRSLPDIDIISGQPKDAWEVQIVKSSWILLCSGLTIG 120
Query 132 LSALAAGLAIGI 143
LS L +G+++GI
Sbjct 121 LSNLFSGISVGI 132
> ath:AT4G32530 vacuolar ATP synthase, putative / V-ATPase, putative;
K03661 V-type H+-transporting ATPase 21kDa proteolipid
subunit [EC:3.6.3.14]
Length=210
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 52/102 (50%), Gaps = 6/102 (5%)
Query 43 FGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIIS 102
F +GI +I + LGAA+G +G + + P + ++++ VI + IYG+I++
Sbjct 25 FSAIGIAISIGVSVLGAAWGIYITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVA 84
Query 103 IVISSVMGSP------DVYSAYAGFGHLAGGLAVGLSALAAG 138
I++ + + S D S AG+ A G+ VG + L G
Sbjct 85 IILQTKLESVPSSKMYDAESLRAGYAIFASGIIVGFANLVCG 126
> cel:T01H3.1 vha-4; Vacuolar H ATPase family member (vha-4);
K03661 V-type H+-transporting ATPase 21kDa proteolipid subunit
[EC:3.6.3.14]
Length=214
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 53/112 (47%), Gaps = 14/112 (12%)
Query 46 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 105
+GI ++ + LGA +G +G + GV P + ++++ +I + I+G+I++ V
Sbjct 54 LGIGFSLSLSVLGAGWGIFTTGSSILGGGVKAPRIRTKNLVSIIFCEAVAIFGIIMAFVF 113
Query 106 SSVMGS------PDVYSAYA--------GFGHLAGGLAVGLSALAAGLAIGI 143
+ PD A G+ GGL VGLS L GLA+GI
Sbjct 114 VGKLAEFRREDLPDTEDGMAILARNLASGYMIFGGGLTVGLSNLVCGLAVGI 165
> pfa:MAL13P1.271 V-type ATPase, putative; K03661 V-type H+-transporting
ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=181
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 56/118 (47%), Gaps = 17/118 (14%)
Query 43 FGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIIS 102
+ +GI ++ + +GAA+G G + V P +I ++++ +I LG+YG+I +
Sbjct 17 WAMLGIALSLFLSIMGAAWGIFICGTSIVGASVKSPRIISKNLISIIFCEALGMYGVITA 76
Query 103 I-----------------VISSVMGSPDVYSAYAGFGHLAGGLAVGLSALAAGLAIGI 143
+ V+++ + + G+ A GL GLS L +G+++GI
Sbjct 77 VFLQIKFSGLSTEVHPPLVLTNKTDPLIMNTIRGGWALFASGLTAGLSNLVSGVSVGI 134
> dre:100329244 solute carrier family 19 (thiamine transporter),
member 2-like; K14610 solute carrier family 19 (thiamine
transporter), member 2/3
Length=478
Score = 32.3 bits (72), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 7 NAHPATTFPHFASSPSLPLFKMFWSAFLQCDPNSVFFGF 45
N P T + +SS L + KM W+ FLQC +S +
Sbjct 243 NKQPNTLKSNTSSSGLLDVLKMLWTDFLQCYSSSTLLAW 281
> dre:569085 solute carrier family 19 (thiamine transporter),
member 2-like; K14610 solute carrier family 19 (thiamine transporter),
member 2/3
Length=458
Score = 32.0 bits (71), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 7 NAHPATTFPHFASSPSLPLFKMFWSAFLQCDPNSVFFGF 45
N P T + +SS L + KM W+ FLQC +S +
Sbjct 223 NKQPNTLKSNTSSSGLLDVLKMLWTDFLQCYSSSTLLAW 261
> tgo:TGME49_091310 vacuolar ATP synthase 22 kDa proteolipid subunit,
putative (EC:3.6.3.14); K03661 V-type H+-transporting
ATPase 21kDa proteolipid subunit [EC:3.6.3.14]
Length=205
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 52/104 (50%), Gaps = 14/104 (13%)
Query 54 FANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVISSVM-GSP 112
F+ +GAA+G G + V P + ++++ VI L IYG+II+I+IS + +P
Sbjct 46 FSVVGAAWGIFICGSSICGAAVRAPRIRSKNLVSVIFCEALAIYGVIIAIIISGQLDNAP 105
Query 113 DVYSA-------------YAGFGHLAGGLAVGLSALAAGLAIGI 143
+S AG+ GL VGLS L G+++G+
Sbjct 106 ANFSPIAGKLTDWQNQAIVAGWALFCCGLTVGLSNLFCGISVGV 149
> dre:568337 olfcd3, VR3.13a, v2r2l2; olfactory receptor C family,
d3; K04613 vomeronasal 2 receptor
Length=813
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 24/61 (39%), Gaps = 9/61 (14%)
Query 4 FFVNAHPATTFPHFASSPSLPLFKMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGT 63
F VN HP P L K FW QC N+ G +G T + ANL Y
Sbjct 303 FLVNVHPD-------HEPKNKLLKEFWETTFQCSFNNR--GSVGCTGSEKLANLQNEYTD 353
Query 64 A 64
A
Sbjct 354 A 354
> dre:567667 slc12a7b, sb:cb734; solute carrier family 12, member
7b; K13627 solute carrier family 12 (potassium/chloride
transporter), member 7
Length=1142
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 28/67 (41%), Gaps = 0/67 (0%)
Query 44 GFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISI 103
G G+TS ++ N+ YG A V V D +P ++ I + L++ I
Sbjct 382 GIPGLTSGVISENMWGKYGPAGMLVEKDIPSVSASDSSQDKYMPYVVNDITAFFTLLVGI 441
Query 104 VISSVMG 110
SV G
Sbjct 442 YFPSVTG 448
Lambda K H
0.327 0.142 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40