bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1345_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
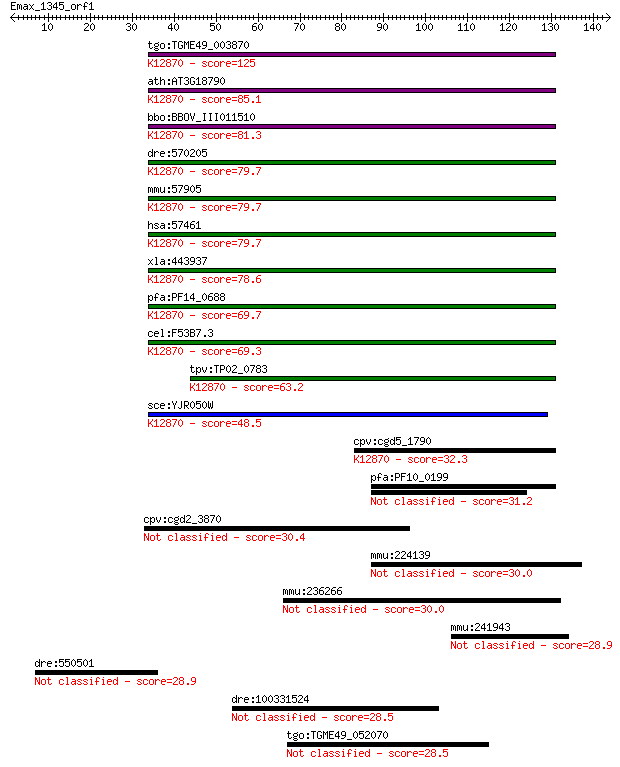
tgo:TGME49_003870 isy1-like splicing family domain-containing ... 125 5e-29
ath:AT3G18790 hypothetical protein; K12870 pre-mRNA-splicing f... 85.1 7e-17
bbo:BBOV_III011510 17.m07982; isy1-like splicing family protei... 81.3 9e-16
dre:570205 rab43, MGC136785, fi21h07, wu:fi21h07, zgc:136785; ... 79.7 3e-15
mmu:57905 Isy1, 5830446M03Rik, AI181014, AU020769; ISY1 splici... 79.7 3e-15
hsa:57461 ISY1, FSAP33, KIAA1160; ISY1 splicing factor homolog... 79.7 3e-15
xla:443937 isy1, MGC80278, fsap33; ISY1 splicing factor homolo... 78.6 7e-15
pfa:PF14_0688 Pre-mRNA-splicing factor ISY1 homolog, putative;... 69.7 3e-12
cel:F53B7.3 hypothetical protein; K12870 pre-mRNA-splicing fac... 69.3 4e-12
tpv:TP02_0783 hypothetical protein; K12870 pre-mRNA-splicing f... 63.2 3e-10
sce:YJR050W ISY1, NTC30, UTR3; Isy1p; K12870 pre-mRNA-splicing... 48.5 6e-06
cpv:cgd5_1790 conserved eukaryotic protein ; K12870 pre-mRNA-s... 32.3 0.54
pfa:PF10_0199 conserved Plasmodium membrane protein 31.2 0.99
cpv:cgd2_3870 ABC transporter with 2x AAA and 11+ transmembran... 30.4 1.7
mmu:224139 Golgb1, 4930428L02Rik, 628101, 6330407A06Rik, AU042... 30.0 2.6
mmu:236266 Alms1, bbb; Alstrom syndrome 1 homolog (human) 30.0 2.6
mmu:241943 Ccdc144b, MGC58791; coiled-coil domain containing 144B 28.9 5.7
dre:550501 wu:fi32a11; zgc:111983 28.9 5.8
dre:100331524 NLR family, pyrin domain containing 1-like 28.5 7.0
tgo:TGME49_052070 hypothetical protein 28.5 7.7
> tgo:TGME49_003870 isy1-like splicing family domain-containing
protein ; K12870 pre-mRNA-splicing factor ISY1
Length=285
Score = 125 bits (314), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 61/97 (62%), Positives = 76/97 (78%), Gaps = 1/97 (1%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAERANAVLNKWLAVK+AV+KGA + P C DL +A +WR +++REIG+
Sbjct 1 MARNAERANAVLNKWLAVKDAVVKGAASR-ERRPRDVNSCQDLKQAEKWRSEVMREIGKL 59
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
I+ +QD+SLG+HRIRDLND IN+ I+ KK WEFRIKE
Sbjct 60 ITQVQDASLGEHRIRDLNDDINRMIRVKKAWEFRIKE 96
> ath:AT3G18790 hypothetical protein; K12870 pre-mRNA-splicing
factor ISY1
Length=300
Score = 85.1 bits (209), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 44/97 (45%), Positives = 66/97 (68%), Gaps = 3/97 (3%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARN E+A ++LN+++ K + K K+ P + EC DL A +WRQQ++REIG
Sbjct 1 MARNEEKAQSMLNRFITQKESEKKKPKER---RPYLASECRDLAEADKWRQQILREIGSK 57
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ+ LG+HR+RDLND+INK ++++ WE RI E
Sbjct 58 VAEIQNEGLGEHRLRDLNDEINKLLRERYHWERRIVE 94
> bbo:BBOV_III011510 17.m07982; isy1-like splicing family protein;
K12870 pre-mRNA-splicing factor ISY1
Length=228
Score = 81.3 bits (199), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 45/99 (45%), Positives = 64/99 (64%), Gaps = 5/99 (5%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQL--GPCIPEECDDLDRATQWRQQLIREIG 91
MARN+E+ANA+LNKWL +K+ + A + QL P E D A WR L++++
Sbjct 1 MARNSEKANAMLNKWLRIKSGL---AAHDTQLTRKPRHTSEVTDYRTAEHWRNLLVKDVM 57
Query 92 RGISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
IS IQ++SLG+ IRDLND+IN+ I +KRW+ R+ E
Sbjct 58 ISISRIQNASLGEFAIRDLNDEINRLIGLRKRWDERVIE 96
> dre:570205 rab43, MGC136785, fi21h07, wu:fi21h07, zgc:136785;
RAB43, member RAS oncogene family; K12870 pre-mRNA-splicing
factor ISY1
Length=285
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 66/97 (68%), Gaps = 4/97 (4%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L ++ + A ++ K + P + EC++L +A +WR+Q+I EI +
Sbjct 1 MARNAEKAMTALARF---RQAQLEEGKVK-ERRPYLASECNELPKAEKWRRQIISEISKK 56
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ++ LG+ +IRDLND+INK +++K WE RIKE
Sbjct 57 VAQIQNAGLGEFKIRDLNDEINKLLREKGHWEVRIKE 93
> mmu:57905 Isy1, 5830446M03Rik, AI181014, AU020769; ISY1 splicing
factor homolog (S. cerevisiae); K12870 pre-mRNA-splicing
factor ISY1
Length=285
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 42/97 (43%), Positives = 65/97 (67%), Gaps = 4/97 (4%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L ++ + A ++ K + P + EC +L +A +WR+Q+I EI +
Sbjct 1 MARNAEKAMTALARF---RQAQLEEGKVK-ERRPFLASECTELPKAEKWRRQIIGEISKK 56
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ++ LG+ RIRDLND+INK +++K WE RIKE
Sbjct 57 VAQIQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKE 93
> hsa:57461 ISY1, FSAP33, KIAA1160; ISY1 splicing factor homolog
(S. cerevisiae); K12870 pre-mRNA-splicing factor ISY1
Length=307
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 42/97 (43%), Positives = 65/97 (67%), Gaps = 4/97 (4%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L ++ + A ++ K + P + EC +L +A +WR+Q+I EI +
Sbjct 1 MARNAEKAMTALARF---RQAQLEEGKVK-ERRPFLASECTELPKAEKWRRQIIGEISKK 56
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ++ LG+ RIRDLND+INK +++K WE RIKE
Sbjct 57 VAQIQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKE 93
> xla:443937 isy1, MGC80278, fsap33; ISY1 splicing factor homolog;
K12870 pre-mRNA-splicing factor ISY1
Length=284
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 65/97 (67%), Gaps = 4/97 (4%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L ++ + A ++ K + P + EC +L +A +WR+Q+I EI +
Sbjct 1 MARNAEKAMTALARF---RQAQLEDGKVK-ERRPFLASECSELPKAEKWRRQIIGEISKK 56
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ++ LG+ +IRD+ND+INK I++K WE RIKE
Sbjct 57 VAQIQNAGLGEFKIRDVNDEINKLIREKGHWEVRIKE 93
> pfa:PF14_0688 Pre-mRNA-splicing factor ISY1 homolog, putative;
K12870 pre-mRNA-splicing factor ISY1
Length=201
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 58/97 (59%), Gaps = 3/97 (3%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARN E+ ++LN+W+ K K+ P +E ++LD A ++R +I+E+ +
Sbjct 1 MARNVEKGKSMLNQWIKAKEI---SDKREFFKIPKNIDEVENLDDALKYRIYIIKEMCKK 57
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
I IQ+ SL D IR+LNDQINK I K +WE RI E
Sbjct 58 IKEIQNHSLSDQHIRELNDQINKLIFIKNKWEARIVE 94
> cel:F53B7.3 hypothetical protein; K12870 pre-mRNA-splicing factor
ISY1
Length=267
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 61/97 (62%), Gaps = 3/97 (3%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L +W +K +G I P ++C +L A ++R++++R+ +
Sbjct 1 MARNAEKAMTALARWRRMKEEEERGP---IARRPHDVKDCRNLSDAERFRREIVRDASKK 57
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
I+ IQ+ LG+ ++RDLND++N+ IK K WE RI+E
Sbjct 58 ITAIQNPGLGEFKLRDLNDEVNRLIKLKHAWEQRIRE 94
> tpv:TP02_0783 hypothetical protein; K12870 pre-mRNA-splicing
factor ISY1
Length=182
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 52/87 (59%), Gaps = 4/87 (4%)
Query 44 VLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRGISLIQDSSLG 103
+LNKWL +K+ G ++ L P E +L A +WR I+EI I+ IQD SLG
Sbjct 1 MLNKWLRIKS----GLEQEQVLRPRHTAEVTNLKEAEKWRSATIKEIMFNINKIQDGSLG 56
Query 104 DHRIRDLNDQINKKIKQKKRWEFRIKE 130
+ +RDLND+IN+ I +K W+ RI E
Sbjct 57 EFVVRDLNDEINRLIGIRKHWDDRIIE 83
> sce:YJR050W ISY1, NTC30, UTR3; Isy1p; K12870 pre-mRNA-splicing
factor ISY1
Length=235
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 54/96 (56%), Gaps = 1/96 (1%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLG-PCIPEECDDLDRATQWRQQLIREIGR 92
M+RN ++AN+VL ++ + G K + P + + A +W++Q+ +EI +
Sbjct 1 MSRNVDKANSVLVRFQEQQAESAGGYKDYSRYQRPRNVSKVKSIKEANEWKRQVSKEIKQ 60
Query 93 GISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRI 128
+ I D SL + +I +LND++N K+ KRW++ I
Sbjct 61 KSTRIYDPSLNEMQIAELNDELNNLFKEWKRWQWHI 96
> cpv:cgd5_1790 conserved eukaryotic protein ; K12870 pre-mRNA-splicing
factor ISY1
Length=175
Score = 32.3 bits (72), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 83 RQQLIREIGRGISLIQDSSLGD-HRIRDLNDQINKKIKQKKRWEFRIKE 130
R +++ EI ++D ++ D + +R LN IN +K+K +WE RI E
Sbjct 5 RFEIMEEISNLAKKLRDCTMSDENEVRHLNSDINSLLKEKYKWECRIVE 53
> pfa:PF10_0199 conserved Plasmodium membrane protein
Length=2134
Score = 31.2 bits (69), Expect = 0.99, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Query 87 IREIGRGISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
IR+ +G I+D+ G+ RIRD N + N++I+ K+ RI++
Sbjct 1755 IRDNKKGNKGIRDNKKGNKRIRD-NKKGNRRIRDNKKGNRRIRD 1797
Score = 28.9 bits (63), Expect = 5.1, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Query 87 IREIGRGISLIQDSSLGDHRIRDLNDQINKKIKQKKR 123
IR+ +G I+D+ G+ RIRD N + N++I+ K+
Sbjct 1765 IRDNKKGNKRIRDNKKGNRRIRD-NKKGNRRIRDNKK 1800
> cpv:cgd2_3870 ABC transporter with 2x AAA and 11+ transmembrane
domains
Length=1928
Score = 30.4 bits (67), Expect = 1.7, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 33/69 (47%), Gaps = 8/69 (11%)
Query 33 KMARNAERANAVLNKWL---AVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWR---QQL 86
K++RN E N+ N W+ K+ ++ G + N ++ C+ + C + W+ L
Sbjct 748 KISRNVEIINS--NPWIPFGTFKDVILAGREYNTEIMECVIQVCQLITDLKMWQLGYNHL 805
Query 87 IREIGRGIS 95
I E G +S
Sbjct 806 IEEYGNNLS 814
> mmu:224139 Golgb1, 4930428L02Rik, 628101, 6330407A06Rik, AU042952,
C130074L01Rik, F730017E11Rik, Gm6840, KIAA4151, MGC183642,
mKIAA4151; golgi autoantigen, golgin subfamily b, macrogolgin
1
Length=3238
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 10/57 (17%)
Query 87 IREIGRGISLIQDSSLGDHRIRDLNDQINKK-------IKQKKRWEFRIKEGDLITQ 136
I+ GR +S +QDS DH +L D + KK + Q K W+ +EGD+++Q
Sbjct 2714 IQAFGRSMSSLQDSR--DHATEELGD-LKKKYDASLKELAQLKEWQDSSREGDVLSQ 2767
> mmu:236266 Alms1, bbb; Alstrom syndrome 1 homolog (human)
Length=3251
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 31/71 (43%), Gaps = 5/71 (7%)
Query 66 GPCIPEE---CDDLDRATQWRQQLIREIGRGISLIQDSSLGDHRIRDLND--QINKKIKQ 120
G C +E + LDR + Q I R QD S G HR R+ Q +K KQ
Sbjct 2711 GACDTKELSLVERLDRLAKLLQNPITHSLRASESAQDDSRGGHRAREWTGRRQQKQKGKQ 2770
Query 121 KKRWEFRIKEG 131
++W ++ G
Sbjct 2771 HRKWSKSLERG 2781
> mmu:241943 Ccdc144b, MGC58791; coiled-coil domain containing
144B
Length=520
Score = 28.9 bits (63), Expect = 5.7, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 106 RIRDLNDQINKKIKQKKRWEFRIKEGDL 133
++R+ DQ NK++K K++ E R++E D+
Sbjct 128 QLREKEDQYNKEVKMKQKLEIRVRELDM 155
> dre:550501 wu:fi32a11; zgc:111983
Length=437
Score = 28.9 bits (63), Expect = 5.8, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 7 STTAATATTTTTAAAAAAAAAAATAAKMA 35
+TTA+TA+ TTA+ A+ A A+TA A
Sbjct 134 ATTASTASGATTASTASGATTASTAGSQA 162
> dre:100331524 NLR family, pyrin domain containing 1-like
Length=959
Score = 28.5 bits (62), Expect = 7.0, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 54 AVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRGISLIQDSSL 102
AVIK K +Q E C+ L A Q ++RE+ I+ +QDS +
Sbjct 588 AVIKFRKALLQWCNLTAESCESLSSALQSSNCVLRELDLSINDLQDSGV 636
> tgo:TGME49_052070 hypothetical protein
Length=292
Score = 28.5 bits (62), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query 67 PCIPEECDDLDRATQWRQQLIREIGRGISLIQD-SSLGDHRIRDLNDQI 114
P I + DD +W+ + + G G+SL D SS G+ R D QI
Sbjct 72 PDIDQPLDDSQNREEWQVEFVATGGPGVSLEGDSSSTGESRGPDSRPQI 120
Lambda K H
0.318 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40