bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1310_orf2
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
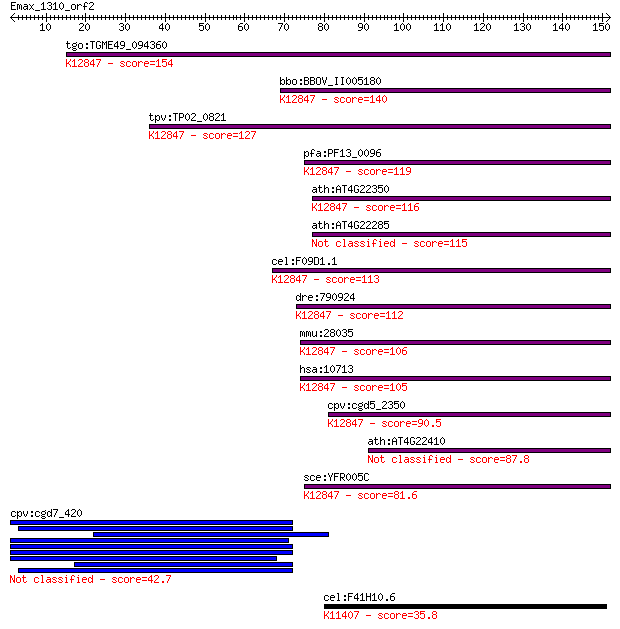
tgo:TGME49_094360 ubiquitin carboxyl-terminal hydrolase, putat... 154 1e-37
bbo:BBOV_II005180 18.m06428; u4/u6.u5 tri-snRNP-associated 65 ... 140 2e-33
tpv:TP02_0821 ubiquitin carboxyl-terminal hydrolase; K12847 U4... 127 1e-29
pfa:PF13_0096 Ubiquitin Carboxyl-terminal Hydrolase-like zinc ... 119 5e-27
ath:AT4G22350 ubiquitin carboxyl-terminal hydrolase family pro... 116 3e-26
ath:AT4G22285 ubiquitin thiolesterase/ zinc ion binding 115 4e-26
cel:F09D1.1 hypothetical protein; K12847 U4/U6.U5 tri-snRNP-as... 113 2e-25
dre:790924 usp39, wu:fb54d10, wu:fb79f05, zgc:158428; ubiquiti... 112 6e-25
mmu:28035 Usp39, AA408960, AI894154, CGI-21, D6Wsu157e, SAD1; ... 106 4e-23
hsa:10713 USP39, HSPC332, MGC75069, SAD1, SNRNP65; ubiquitin s... 105 4e-23
cpv:cgd5_2350 SnRNP assembly defective 1 like ubiquitin C-term... 90.5 2e-18
ath:AT4G22410 ubiquitin carboxyl-terminal hydrolase family pro... 87.8 1e-17
sce:YFR005C SAD1; Conserved zinc-finger domain protein involve... 81.6 9e-16
cpv:cgd7_420 protein with DEXDc plus ring plus HELICc; possibl... 42.7 4e-04
cel:F41H10.6 hypothetical protein; K11407 histone deacetylase ... 35.8 0.058
> tgo:TGME49_094360 ubiquitin carboxyl-terminal hydrolase, putative
(EC:4.1.1.70 3.1.2.15); K12847 U4/U6.U5 tri-snRNP-associated
protein 2
Length=571
Score = 154 bits (389), Expect = 1e-37, Method: Composition-based stats.
Identities = 70/140 (50%), Positives = 100/140 (71%), Gaps = 3/140 (2%)
Query 15 SVGSKRPNEAS-SDVKSSEDSSSNDSSSSATNNSSTTSDNTNS--NTSSSNNGSSSSSSA 71
S S P S +D +S +S + ++ S+ ++ + + + + S+++ SA
Sbjct 6 STASPPPTRDSMADSPASSESPAGPQKRASLQEGSSLAEAAGAAKKRTPAPSPSAAAVSA 65
Query 72 PVQKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMH 131
P K++RRTCPYLGTI +H+LDFDFEKVC ICLSNQHVYACLVC RYFQGRG++T+A+MH
Sbjct 66 PQTKQLRRTCPYLGTIKRHLLDFDFEKVCCICLSNQHVYACLVCARYFQGRGKNTYAYMH 125
Query 132 ALEQRHFVYVNLTTCKVYCL 151
AL+++H+VY+NL C+VYCL
Sbjct 126 ALQEQHYVYLNLKDCRVYCL 145
> bbo:BBOV_II005180 18.m06428; u4/u6.u5 tri-snRNP-associated 65
kDa protein; K12847 U4/U6.U5 tri-snRNP-associated protein
2
Length=470
Score = 140 bits (353), Expect = 2e-33, Method: Composition-based stats.
Identities = 58/83 (69%), Positives = 72/83 (86%), Gaps = 2/83 (2%)
Query 69 SSAPVQKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFA 128
S P +K V+ CPYLGTIN+H+LDFDFEKVCSI LSN+HVYACLVCGRYF+GRG++T+A
Sbjct 30 SEIPAKKHVK--CPYLGTINRHLLDFDFEKVCSITLSNKHVYACLVCGRYFEGRGKNTYA 87
Query 129 FMHALEQRHFVYVNLTTCKVYCL 151
+ HALE+RH+V++NL CKVY L
Sbjct 88 YTHALEERHYVFINLHDCKVYSL 110
> tpv:TP02_0821 ubiquitin carboxyl-terminal hydrolase; K12847
U4/U6.U5 tri-snRNP-associated protein 2
Length=537
Score = 127 bits (319), Expect = 1e-29, Method: Composition-based stats.
Identities = 61/120 (50%), Positives = 81/120 (67%), Gaps = 5/120 (4%)
Query 36 SNDSSSSATNNSSTTSDNTNSNTSSSNNGSSSSS----SAPVQKRVRRTCPYLGTINKHM 91
+ D+ S NN S T + N + S S P + +V CPYLGTIN+H+
Sbjct 96 NEDNELSENNNKSLDETLTIAQCEEPLNDQNESPVPKVSDPPKVKVL-NCPYLGTINRHL 154
Query 92 LDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHFVYVNLTTCKVYCL 151
LDFDFEKVCSI LSN HVYACLVCG+YFQGRG++T+ + HALE+ H++++NL C+VYC+
Sbjct 155 LDFDFEKVCSITLSNIHVYACLVCGKYFQGRGKNTYCYTHALEECHYLFMNLEDCRVYCI 214
> pfa:PF13_0096 Ubiquitin Carboxyl-terminal Hydrolase-like zinc
finger protein; K12847 U4/U6.U5 tri-snRNP-associated protein
2
Length=640
Score = 119 bits (297), Expect = 5e-27, Method: Composition-based stats.
Identities = 49/77 (63%), Positives = 62/77 (80%), Gaps = 0/77 (0%)
Query 75 KRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALE 134
K+ R CPYL TIN+++LDFDFEK+CSI LSN HVYACLVCG YFQG G+ T A+ H+LE
Sbjct 82 KKRNRVCPYLRTINRNLLDFDFEKLCSISLSNLHVYACLVCGLYFQGIGKGTHAYTHSLE 141
Query 135 QRHFVYVNLTTCKVYCL 151
+ H+V++NL TCK C+
Sbjct 142 KNHYVFINLETCKTCCI 158
> ath:AT4G22350 ubiquitin carboxyl-terminal hydrolase family protein;
K12847 U4/U6.U5 tri-snRNP-associated protein 2
Length=510
Score = 116 bits (291), Expect = 3e-26, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 77 VRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQR 136
VRR CPYL T+N+ +LDFDFE+ CS+ LSN +VYACLVCG+YFQGR + + A+ H+LE
Sbjct 76 VRRDCPYLDTVNRQVLDFDFERFCSVSLSNLNVYACLVCGKYFQGRSQKSHAYTHSLEAG 135
Query 137 HFVYVNLTTCKVYCL 151
H VY+NL T KVYCL
Sbjct 136 HHVYINLLTEKVYCL 150
> ath:AT4G22285 ubiquitin thiolesterase/ zinc ion binding
Length=541
Score = 115 bits (289), Expect = 4e-26, Method: Composition-based stats.
Identities = 48/75 (64%), Positives = 60/75 (80%), Gaps = 0/75 (0%)
Query 77 VRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQR 136
VRR CPYL T+N+ +LDFDFE+ CS+ LSN +VYACLVCG+YFQGR + + A+ H+LE
Sbjct 107 VRRDCPYLDTVNRQVLDFDFERFCSVSLSNLNVYACLVCGKYFQGRSQKSHAYTHSLEAG 166
Query 137 HFVYVNLTTCKVYCL 151
H VY+NL T KVYCL
Sbjct 167 HHVYINLLTEKVYCL 181
> cel:F09D1.1 hypothetical protein; K12847 U4/U6.U5 tri-snRNP-associated
protein 2
Length=602
Score = 113 bits (283), Expect = 2e-25, Method: Composition-based stats.
Identities = 50/85 (58%), Positives = 64/85 (75%), Gaps = 1/85 (1%)
Query 67 SSSSAPVQKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRST 126
S A +K+ R CPYL TI++ +LDFDFEK CS+ LS+Q+VYAC+VCG+YFQGRG +T
Sbjct 123 SMKKAQAEKK-SRMCPYLDTIDRSVLDFDFEKQCSVSLSHQNVYACMVCGKYFQGRGTNT 181
Query 127 FAFMHALEQRHFVYVNLTTCKVYCL 151
A+ HALE H V++NL T K YCL
Sbjct 182 HAYTHALETDHHVFLNLQTLKFYCL 206
> dre:790924 usp39, wu:fb54d10, wu:fb79f05, zgc:158428; ubiquitin
specific peptidase 39; K12847 U4/U6.U5 tri-snRNP-associated
protein 2
Length=497
Score = 112 bits (279), Expect = 6e-25, Method: Composition-based stats.
Identities = 46/79 (58%), Positives = 63/79 (79%), Gaps = 0/79 (0%)
Query 73 VQKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHA 132
V+ R R CPYL TIN+ +LDFDFEK+CSI LS+ +VYACL+CG+YFQGRG+ + A++H+
Sbjct 28 VEDRRSRHCPYLDTINRSVLDFDFEKLCSISLSHINVYACLICGKYFQGRGQKSHAYIHS 87
Query 133 LEQRHFVYVNLTTCKVYCL 151
++ H V++NL T K YCL
Sbjct 88 VQVAHHVFLNLHTLKFYCL 106
> mmu:28035 Usp39, AA408960, AI894154, CGI-21, D6Wsu157e, SAD1;
ubiquitin specific peptidase 39; K12847 U4/U6.U5 tri-snRNP-associated
protein 2
Length=564
Score = 106 bits (264), Expect = 4e-23, Method: Composition-based stats.
Identities = 45/78 (57%), Positives = 60/78 (76%), Gaps = 0/78 (0%)
Query 74 QKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHAL 133
+ R R CPYL TIN+ +LDFDFEK+CSI LS+ + YACLVCG+YFQGRG + A++H++
Sbjct 97 EDRRSRHCPYLDTINRSVLDFDFEKLCSISLSHINAYACLVCGKYFQGRGLKSHAYIHSV 156
Query 134 EQRHFVYVNLTTCKVYCL 151
+ H V++NL T K YCL
Sbjct 157 QFSHHVFLNLHTLKFYCL 174
> hsa:10713 USP39, HSPC332, MGC75069, SAD1, SNRNP65; ubiquitin
specific peptidase 39; K12847 U4/U6.U5 tri-snRNP-associated
protein 2
Length=565
Score = 105 bits (263), Expect = 4e-23, Method: Composition-based stats.
Identities = 45/78 (57%), Positives = 60/78 (76%), Gaps = 0/78 (0%)
Query 74 QKRVRRTCPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHAL 133
+ R R CPYL TIN+ +LDFDFEK+CSI LS+ + YACLVCG+YFQGRG + A++H++
Sbjct 98 EDRRSRHCPYLDTINRSVLDFDFEKLCSISLSHINAYACLVCGKYFQGRGLKSHAYIHSV 157
Query 134 EQRHFVYVNLTTCKVYCL 151
+ H V++NL T K YCL
Sbjct 158 QFSHHVFLNLHTLKFYCL 175
> cpv:cgd5_2350 SnRNP assembly defective 1 like ubiquitin C-terminal
hydrolase with a UBP finger at the N-terminus ; K12847
U4/U6.U5 tri-snRNP-associated protein 2
Length=498
Score = 90.5 bits (223), Expect = 2e-18, Method: Composition-based stats.
Identities = 34/71 (47%), Positives = 53/71 (74%), Gaps = 0/71 (0%)
Query 81 CPYLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHFVY 140
CPYL +I + +LDFD+EK+CS+ L +H+Y CLVCG +QG+G+ + A+ H+LE +H ++
Sbjct 12 CPYLSSIKRSVLDFDYEKICSVTLREEHIYCCLVCGINYQGKGKGSMAYKHSLELKHHLF 71
Query 141 VNLTTCKVYCL 151
+NLT + CL
Sbjct 72 INLTNSSIICL 82
> ath:AT4G22410 ubiquitin carboxyl-terminal hydrolase family protein
Length=340
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 38/61 (62%), Positives = 48/61 (78%), Gaps = 0/61 (0%)
Query 91 MLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHFVYVNLTTCKVYC 150
+LDF FE+ CS+ LSN +VYACLVCG+YFQGR + + A+ H+LE H VY+NL T KVYC
Sbjct 6 VLDFHFERFCSVSLSNLNVYACLVCGKYFQGRSQKSHAYTHSLEAGHHVYINLLTEKVYC 65
Query 151 L 151
L
Sbjct 66 L 66
> sce:YFR005C SAD1; Conserved zinc-finger domain protein involved
in pre-mRNA splicing, required for assembly of U4 snRNA
into the U4/U6 particle; K12847 U4/U6.U5 tri-snRNP-associated
protein 2
Length=448
Score = 81.6 bits (200), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 37/80 (46%), Positives = 54/80 (67%), Gaps = 3/80 (3%)
Query 75 KRVRRTCP---YLGTINKHMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMH 131
K+++ P YL T+ + LDFD EK+C I LS +VY CLVCG Y+QGR + AF+H
Sbjct 20 KKIKSQEPNYAYLETVVREKLDFDSEKICCITLSPLNVYCCLVCGHYYQGRHEKSPAFIH 79
Query 132 ALEQRHFVYVNLTTCKVYCL 151
++++ H V++NLT+ K Y L
Sbjct 80 SIDENHHVFLNLTSLKFYML 99
> cpv:cgd7_420 protein with DEXDc plus ring plus HELICc; possible
SNF2 domain
Length=2042
Score = 42.7 bits (99), Expect = 4e-04, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 44/71 (61%), Gaps = 0/71 (0%)
Query 1 AETAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSS 60
+ TA + A + + + N A+S+ +S S+SN ++S+ +N+++ S+ NSNTS+
Sbjct 289 SNTANSNTANSNTANFNTANSNTANSNTANSNTSNSNTANSNTSNSNTANSNTANSNTSN 348
Query 61 SNNGSSSSSSA 71
SN +S ++++
Sbjct 349 SNTANSITTTS 359
Score = 38.5 bits (88), Expect = 0.008, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 39/59 (66%), Gaps = 0/59 (0%)
Query 22 NEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSSSNNGSSSSSSAPVQKRVRRT 80
N A+ + +S ++SN ++S+ +N+++ S+ +NSNT++SN +S++S++ + T
Sbjct 300 NTANFNTANSNTANSNTANSNTSNSNTANSNTSNSNTANSNTANSNTSNSNTANSITTT 358
Score = 38.5 bits (88), Expect = 0.009, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 42/69 (60%), Gaps = 0/69 (0%)
Query 3 TAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSSSN 62
TA + A + + N A+S+ + ++SN ++S+ N++++ S+ NSNTS+SN
Sbjct 276 TANSNTANFNTTNSNTANSNTANSNTANFNTANSNTANSNTANSNTSNSNTANSNTSNSN 335
Query 63 NGSSSSSSA 71
+S+++++
Sbjct 336 TANSNTANS 344
Score = 38.1 bits (87), Expect = 0.009, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 41/70 (58%), Gaps = 0/70 (0%)
Query 1 AETAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSS 60
+ TA + A + + N A+S+ +S ++SN S+S+ N+++ S+ +NSNT++
Sbjct 294 SNTANSNTANFNTANSNTANSNTANSNTSNSNTANSNTSNSNTANSNTANSNTSNSNTAN 353
Query 61 SNNGSSSSSS 70
S +SS+ S
Sbjct 354 SITTTSSNCS 363
Score = 37.4 bits (85), Expect = 0.017, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 1 AETAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSS 60
+ TA A + + + N A+S+ +S ++ N ++S+ N+++ S+ +NSNT++
Sbjct 269 SNTANFNTANSNTANFNTTNSNTANSNTANSNTANFNTANSNTANSNTANSNTSNSNTAN 328
Query 61 SNNGSSSSSSA 71
SN +S+++++
Sbjct 329 SNTSNSNTANS 339
Score = 35.4 bits (80), Expect = 0.064, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 1 AETAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSS 60
+ TA A + + + N A+ + +S ++SN ++S+ N ++ S+ NSNT++
Sbjct 259 SNTANFNTANSNTANFNTANSNTANFNTTNSNTANSNTANSNTANFNTANSNTANSNTAN 318
Query 61 SNNGSSSSSSA 71
SN +S+++++
Sbjct 319 SNTSNSNTANS 329
Score = 35.0 bits (79), Expect = 0.090, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 39/67 (58%), Gaps = 0/67 (0%)
Query 1 AETAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSS 60
+ TA A + + + N ++S+ +S S+SN ++S+ N++++ S+ NS T++
Sbjct 299 SNTANFNTANSNTANSNTANSNTSNSNTANSNTSNSNTANSNTANSNTSNSNTANSITTT 358
Query 61 SNNGSSS 67
S+N S S
Sbjct 359 SSNCSRS 365
Score = 33.5 bits (75), Expect = 0.28, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 17 GSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSSSNNGSSSSSSA 71
G+ N A+ + +S ++ N ++S+ N ++T S+ NSNT++SN + +++++
Sbjct 255 GTANSNTANFNTANSNTANFNTANSNTANFNTTNSNTANSNTANSNTANFNTANS 309
Score = 32.7 bits (73), Expect = 0.48, Method: Composition-based stats.
Identities = 16/69 (23%), Positives = 39/69 (56%), Gaps = 0/69 (0%)
Query 3 TAAAAAAAAAADSVGSKRPNEASSDVKSSEDSSSNDSSSSATNNSSTTSDNTNSNTSSSN 62
TA A + + + N A+ + +S ++ N ++S+ N+++ S+ N NT++SN
Sbjct 251 TANFGTANSNTANFNTANSNTANFNTANSNTANFNTTNSNTANSNTANSNTANFNTANSN 310
Query 63 NGSSSSSSA 71
+S+++++
Sbjct 311 TANSNTANS 319
> cel:F41H10.6 hypothetical protein; K11407 histone deacetylase
6/10 [EC:3.5.1.98]
Length=955
Score = 35.8 bits (81), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 1/72 (1%)
Query 80 TCPYLGTINK-HMLDFDFEKVCSICLSNQHVYACLVCGRYFQGRGRSTFAFMHALEQRHF 138
TCP+L + + CS C V+ CL C +Y GR + A MH L H
Sbjct 854 TCPHLKEVKPLPPAKINARTACSECQIGAEVWTCLTCYKYNCGRFVNEHAMMHHLSSSHP 913
Query 139 VYVNLTTCKVYC 150
+ +++ V+C
Sbjct 914 MALSMADLSVWC 925
Lambda K H
0.311 0.119 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40