bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1292_orf1
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
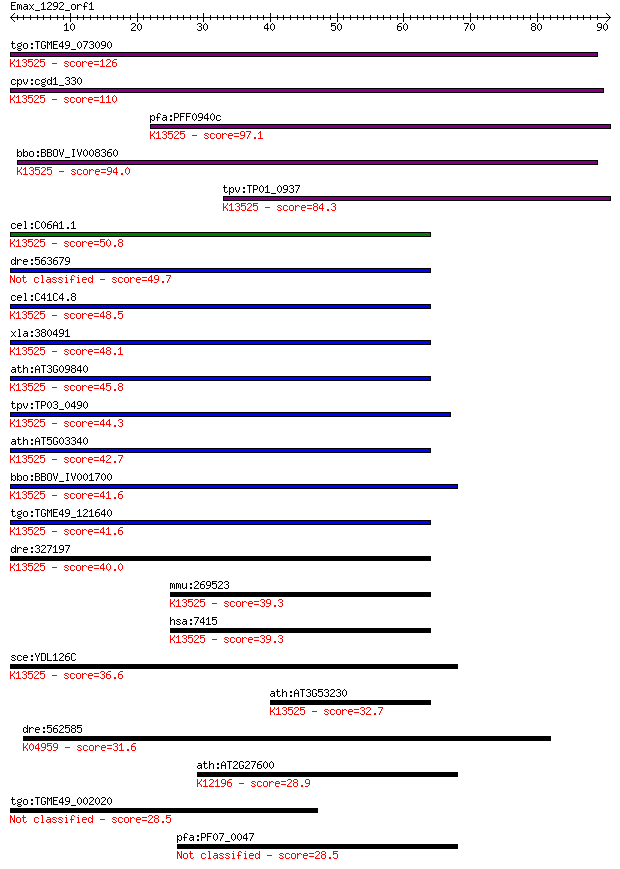
tgo:TGME49_073090 cell division protein 48, putative (EC:3.4.2... 126 1e-29
cpv:cgd1_330 CDC48 like AAA ATPase ortholog ; K13525 transitio... 110 1e-24
pfa:PFF0940c cell division cycle protein 48 homologue, putativ... 97.1 1e-20
bbo:BBOV_IV008360 23.m05756; cell division control protein 48;... 94.0 1e-19
tpv:TP01_0937 cell division cycle protein 48; K13525 transitio... 84.3 9e-17
cel:C06A1.1 cdc-48.1; Cell Division Cycle related family membe... 50.8 1e-06
dre:563679 MGC136908; zgc:136908 49.7 2e-06
cel:C41C4.8 cdc-48.2; Cell Division Cycle related family membe... 48.5 5e-06
xla:380491 vcp, MGC52611; valosin containing protein; K13525 t... 48.1 6e-06
ath:AT3G09840 CDC48; CDC48 (CELL DIVISION CYCLE 48); ATPase/ i... 45.8 3e-05
tpv:TP03_0490 cell division cycle protein 48; K13525 transitio... 44.3 1e-04
ath:AT5G03340 cell division cycle protein 48, putative / CDC48... 42.7 3e-04
bbo:BBOV_IV001700 21.m02769; cell division cycle protein ATPas... 41.6 6e-04
tgo:TGME49_121640 cell division protein 48, putative ; K13525 ... 41.6 6e-04
dre:327197 vcp, CDC48, wu:fd16d05, wu:fj63d11; valosin contain... 40.0 0.002
mmu:269523 Vcp, 3110001E05, CDC48, p97, p97/VCP; valosin conta... 39.3 0.003
hsa:7415 VCP, IBMPFD, MGC131997, MGC148092, MGC8560, TERA, p97... 39.3 0.003
sce:YDL126C CDC48; ATPase in ER, nuclear membrane and cytosol ... 36.6 0.022
ath:AT3G53230 cell division cycle protein 48, putative / CDC48... 32.7 0.25
dre:562585 itpr2, si:dkey-196d8.1; inositol 1,4,5-triphosphate... 31.6 0.59
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 28.9 4.6
tgo:TGME49_002020 hypothetical protein 28.5 4.8
pfa:PF07_0047 cell division cycle ATPase, putative 28.5 5.2
> tgo:TGME49_073090 cell division protein 48, putative (EC:3.4.21.53);
K13525 transitional endoplasmic reticulum ATPase
Length=811
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 64/94 (68%), Positives = 79/94 (84%), Gaps = 8/94 (8%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGMDED------ESNVKYEITRKHFEEGIAGARRSVSQ 54
RAAKAAIRDAIAAEELA+ N G DE ++++ YEITRKHFEEG+AGARRSVSQ
Sbjct 699 RAAKAAIRDAIAAEELAQV--NAGADEMDAEEEEKTDIVYEITRKHFEEGLAGARRSVSQ 756
Query 55 TDLSKYDSFRMKFDPIYKSQAAGGDGTIIVDWPN 88
TDL+KYD+FRMKFDP+YKSQAAGG+ ++++WP+
Sbjct 757 TDLTKYDNFRMKFDPLYKSQAAGGETQVLIEWPD 790
> cpv:cgd1_330 CDC48 like AAA ATPase ortholog ; K13525 transitional
endoplasmic reticulum ATPase
Length=820
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/93 (63%), Positives = 74/93 (79%), Gaps = 7/93 (7%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGM----DEDESNVKYEITRKHFEEGIAGARRSVSQTD 56
RAAKAAIRDAIAAEEL +A+G++ DE +S++ YEI RKHFEE AGARRSVS TD
Sbjct 713 RAAKAAIRDAIAAEELKKASGDDSAMKIEDEVDSHI-YEIGRKHFEEAFAGARRSVSITD 771
Query 57 LSKYDSFRMKFDPIYKSQAAGGDGTIIVDWPNT 89
L+KYD FRMKFDP+Y +Q +GG+G +DWP++
Sbjct 772 LAKYDQFRMKFDPVYVTQ-SGGEG-FTIDWPDS 802
> pfa:PFF0940c cell division cycle protein 48 homologue, putative;
K13525 transitional endoplasmic reticulum ATPase
Length=828
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 41/69 (59%), Positives = 53/69 (76%), Gaps = 0/69 (0%)
Query 22 NEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKFDPIYKSQAAGGDGT 81
N+ D+ N+KYEITR HF+EG+AGARRSVSQ DL KYD+FR+KFDP+YK++ G
Sbjct 744 NDQQKNDDDNIKYEITRHHFKEGLAGARRSVSQADLIKYDNFRIKFDPLYKTKTGGTGDD 803
Query 82 IIVDWPNTD 90
I+DWP+ D
Sbjct 804 FIIDWPDED 812
> bbo:BBOV_IV008360 23.m05756; cell division control protein 48;
K13525 transitional endoplasmic reticulum ATPase
Length=804
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 48/87 (55%), Positives = 62/87 (71%), Gaps = 5/87 (5%)
Query 2 AAKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYD 61
AA++AIRDAIA EE EG + YEI RKHF+EG+A AR SV+ TDL+K+D
Sbjct 704 AARSAIRDAIAYEEKHGKTPTEGT----PDFTYEIQRKHFQEGLANARHSVTSTDLAKFD 759
Query 62 SFRMKFDPIYKSQAAGGDGTIIVDWPN 88
+FR KFDP+YK++ AGGD I +DWP+
Sbjct 760 NFRNKFDPLYKTRGAGGD-EIDIDWPD 785
> tpv:TP01_0937 cell division cycle protein 48; K13525 transitional
endoplasmic reticulum ATPase
Length=811
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 34/58 (58%), Positives = 46/58 (79%), Gaps = 0/58 (0%)
Query 33 KYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKFDPIYKSQAAGGDGTIIVDWPNTD 90
KYEITRKHF+EG+A AR SV+ +D++KYD+FR KFDP+YK++ + G I +DWP D
Sbjct 740 KYEITRKHFQEGLANARHSVTSSDITKYDAFRTKFDPLYKNRNSTGQNDIDIDWPEND 797
> cel:C06A1.1 cdc-48.1; Cell Division Cycle related family member
(cdc-48.1); K13525 transitional endoplasmic reticulum ATPase
Length=809
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 41/70 (58%), Gaps = 7/70 (10%)
Query 1 RAAKAAIRDAIAAE-------ELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVS 53
RA K AIR++I E + +A G E M++D + EITR HFEE + ARRSV+
Sbjct 700 RACKLAIRESIEKEIRIEKERQDRQARGEELMEDDAVDPVPEITRAHFEEAMKFARRSVT 759
Query 54 QTDLSKYDSF 63
D+ KY+ F
Sbjct 760 DNDIRKYEMF 769
> dre:563679 MGC136908; zgc:136908
Length=805
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDL 57
RA K AIR+AI AE E R A E +D+ + EI + HFEE + ARRSVS D+
Sbjct 695 RACKLAIREAIEAEIRAERQRQARKETAMDDDYDPVPEIRKDHFEEAMRFARRSVSDNDI 754
Query 58 SKYDSF 63
KY+ F
Sbjct 755 RKYEMF 760
> cel:C41C4.8 cdc-48.2; Cell Division Cycle related family member
(cdc-48.2); K13525 transitional endoplasmic reticulum ATPase
Length=810
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 42/70 (60%), Gaps = 7/70 (10%)
Query 1 RAAKAAIRDAIAAE---ELAR----AAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVS 53
RA K AIR++I E E R A G E M+++ ++ EITR HFEE + ARRSV+
Sbjct 698 RACKLAIRESIEREIRQEKERQDRSARGEELMEDELADPVPEITRAHFEEAMKFARRSVT 757
Query 54 QTDLSKYDSF 63
D+ KY+ F
Sbjct 758 DNDIRKYEMF 767
> xla:380491 vcp, MGC52611; valosin containing protein; K13525
transitional endoplasmic reticulum ATPase
Length=805
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 37/66 (56%), Gaps = 3/66 (4%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDL 57
RA K AIR++I E E R M+ +E + EI R HFEE + ARRSVS D+
Sbjct 693 RACKLAIRESIENEIRRERDRQTNPSAMEVEEDDPVPEIRRDHFEEAMRLARRSVSDNDI 752
Query 58 SKYDSF 63
KY+ F
Sbjct 753 RKYEMF 758
> ath:AT3G09840 CDC48; CDC48 (CELL DIVISION CYCLE 48); ATPase/
identical protein binding; K13525 transitional endoplasmic
reticulum ATPase
Length=809
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 37/66 (56%), Gaps = 3/66 (4%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDL 57
RA K AIR+ I + E R+ E M+ED + EI HFEE + ARRSVS D+
Sbjct 697 RACKYAIRENIEKDIEKEKRRSENPEAMEEDGVDEVSEIKAAHFEESMKYARRSVSDADI 756
Query 58 SKYDSF 63
KY +F
Sbjct 757 RKYQAF 762
> tpv:TP03_0490 cell division cycle protein 48; K13525 transitional
endoplasmic reticulum ATPase
Length=954
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 41/66 (62%), Gaps = 4/66 (6%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKY 60
RAA+ AIR++I EE+ R +++ E + IT KHF+ + +R+SV Q+D+ Y
Sbjct 890 RAAREAIRESIE-EEIKR---KRPLEKGEKDPVPFITNKHFQVALRNSRKSVEQSDIQLY 945
Query 61 DSFRMK 66
+SF+ K
Sbjct 946 ESFKNK 951
> ath:AT5G03340 cell division cycle protein 48, putative / CDC48,
putative; K13525 transitional endoplasmic reticulum ATPase
Length=810
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 38/67 (56%), Gaps = 4/67 (5%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMDEDESNVKY-EITRKHFEEGIAGARRSVSQTD 56
RA K AIR+ I + E R+ E M+ED + + EI HFEE + ARRSVS D
Sbjct 696 RACKYAIRENIEKDIENERRRSQNPEAMEEDMVDDEVSEIRAAHFEESMKYARRSVSDAD 755
Query 57 LSKYDSF 63
+ KY +F
Sbjct 756 IRKYQAF 762
> bbo:BBOV_IV001700 21.m02769; cell division cycle protein ATPase;
K13525 transitional endoplasmic reticulum ATPase
Length=922
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 40/67 (59%), Gaps = 4/67 (5%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKY 60
RAA+ AIR++I E+ R G + +E V Y IT +HF +A AR+SV + D+ +Y
Sbjct 855 RAAREAIRESIE-HEIKR--GRRLKEGEEDPVPY-ITNEHFRVAMANARKSVRKEDIKRY 910
Query 61 DSFRMKF 67
+ F+ K
Sbjct 911 EQFKKKL 917
> tgo:TGME49_121640 cell division protein 48, putative ; K13525
transitional endoplasmic reticulum ATPase
Length=963
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 35/63 (55%), Gaps = 4/63 (6%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKY 60
RAAK A+R++I AE A + E E + I++KHF+E GARRSV + + Y
Sbjct 863 RAAKNAVRESIQAE----VARGRPLAEGEKDPVPFISKKHFDEAFKGARRSVPEDMVKVY 918
Query 61 DSF 63
F
Sbjct 919 TQF 921
> dre:327197 vcp, CDC48, wu:fd16d05, wu:fj63d11; valosin containing
protein; K13525 transitional endoplasmic reticulum ATPase
Length=806
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 35/66 (53%), Gaps = 3/66 (4%)
Query 1 RAAKAAIRDAIAAEELARA---AGNEGMDEDESNVKYEITRKHFEEGIAGARRSVSQTDL 57
RA K AIR++I E M+ +E + EI + HFEE + ARRSVS D+
Sbjct 693 RACKLAIRESIENEIRRERERQTNPSAMEVEEDDPVPEIRKDHFEEAMRFARRSVSDNDI 752
Query 58 SKYDSF 63
KY+ F
Sbjct 753 RKYEMF 758
> mmu:269523 Vcp, 3110001E05, CDC48, p97, p97/VCP; valosin containing
protein; K13525 transitional endoplasmic reticulum ATPase
Length=806
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 25 MDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSF 63
M+ +E + EI R HFEE + ARRSVS D+ KY+ F
Sbjct 720 MEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKYEMF 758
> hsa:7415 VCP, IBMPFD, MGC131997, MGC148092, MGC8560, TERA, p97;
valosin containing protein; K13525 transitional endoplasmic
reticulum ATPase
Length=806
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 25 MDEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSF 63
M+ +E + EI R HFEE + ARRSVS D+ KY+ F
Sbjct 720 MEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKYEMF 758
> sce:YDL126C CDC48; ATPase in ER, nuclear membrane and cytosol
with homology to mammalian p97; in a complex with Npl4p and
Ufd1p participates in retrotranslocation of ubiquitinated
proteins from the ER into the cytosol for degradation by the
proteasome; K13525 transitional endoplasmic reticulum ATPase
Length=835
Score = 36.6 bits (83), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 41/82 (50%), Gaps = 15/82 (18%)
Query 1 RAAKAAIRDAIAAE---ELARAAGNEGMD------------EDESNVKYEITRKHFEEGI 45
RAAK AI+D+I A E + EG D E E + IT++HF E +
Sbjct 703 RAAKYAIKDSIEAHRQHEAEKEVKVEGEDVEMTDEGAKAEQEPEVDPVPYITKEHFAEAM 762
Query 46 AGARRSVSQTDLSKYDSFRMKF 67
A+RSVS +L +Y+++ +
Sbjct 763 KTAKRSVSDAELRRYEAYSQQM 784
> ath:AT3G53230 cell division cycle protein 48, putative / CDC48,
putative; K13525 transitional endoplasmic reticulum ATPase
Length=815
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 40 HFEEGIAGARRSVSQTDLSKYDSF 63
HFEE + ARRSVS D+ KY +F
Sbjct 738 HFEESMKYARRSVSDADIRKYQAF 761
> dre:562585 itpr2, si:dkey-196d8.1; inositol 1,4,5-triphosphate
receptor, type 2; K04959 inositol 1,4,5-triphosphate receptor
type 2
Length=2588
Score = 31.6 bits (70), Expect = 0.59, Method: Composition-based stats.
Identities = 17/81 (20%), Positives = 35/81 (43%), Gaps = 2/81 (2%)
Query 3 AKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRK--HFEEGIAGARRSVSQTDLSKY 60
A+ IR +++ + + + S ++++ R H E + G + S Y
Sbjct 1801 AQKEIRSSVSVNMFELSCRKREEESEGSGIRHKKVRDSLHLREEMRGQLKDASSVTSKAY 1860
Query 61 DSFRMKFDPIYKSQAAGGDGT 81
+FR ++DP ++ GD T
Sbjct 1861 STFRREWDPDIEAVGTTGDAT 1881
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 29 ESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKF 67
E + ITR FE+ +A R +VS++DL ++ F +F
Sbjct 393 EKIIPPPITRTDFEKVLARQRPTVSKSDLDVHERFTQEF 431
> tgo:TGME49_002020 hypothetical protein
Length=763
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 1 RAAKAAIRDAIAAEELARAAGNEGMDEDESNVKYEITRKHFEEGIA 46
R +KA ++ A ++ +AA + S+V+Y+ITRKHF++ A
Sbjct 332 RMSKACMKAIRALCDVNQAAIDLKFILKNSDVRYDITRKHFDKLCA 377
> pfa:PF07_0047 cell division cycle ATPase, putative
Length=1229
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 26 DEDESNVKYEITRKHFEEGIAGARRSVSQTDLSKYDSFRMKF 67
D D + +++KHF+ AR S+ D+ KY+ F+ K
Sbjct 1183 DTDTYDPVPTLSKKHFDLAFKNARISIQPEDVLKYEKFKEKL 1224
Lambda K H
0.312 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012433808
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40