bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1267_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
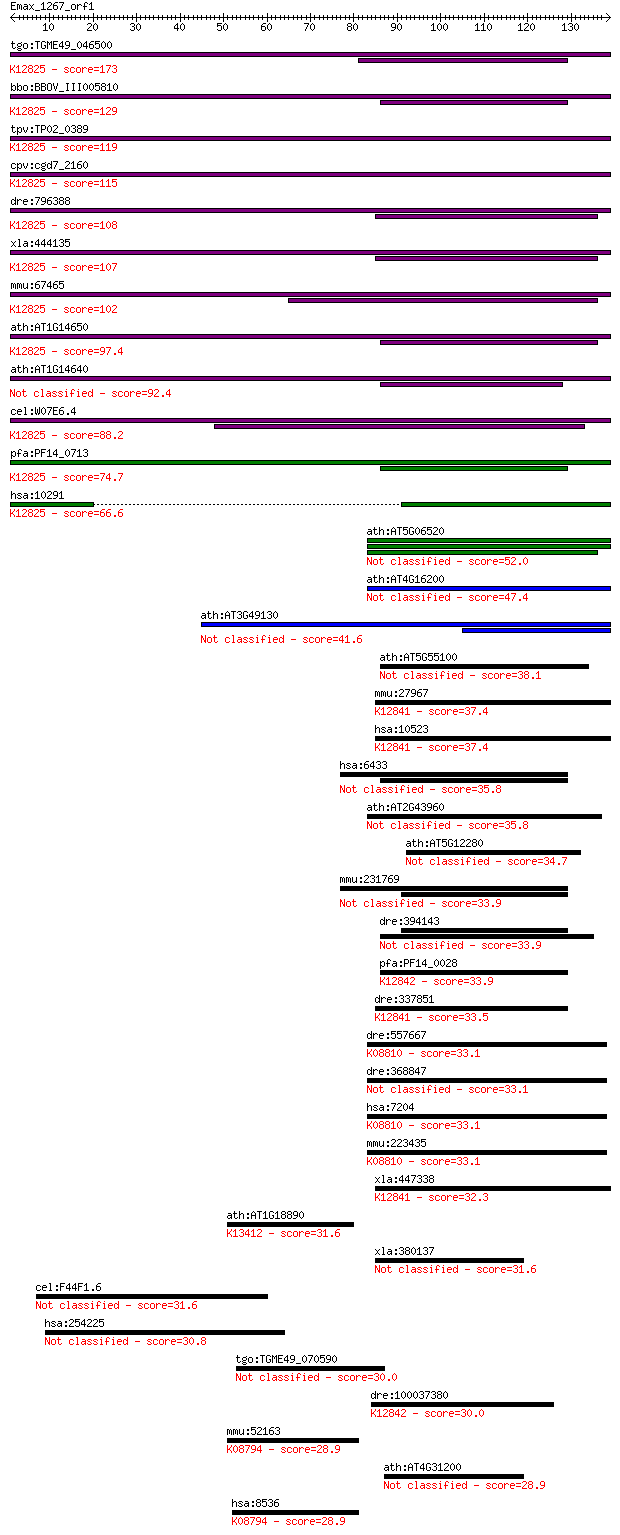
tgo:TGME49_046500 surp module domain-containing protein ; K128... 173 2e-43
bbo:BBOV_III005810 17.m07515; Surp module domain containing pr... 129 3e-30
tpv:TP02_0389 hypothetical protein; K12825 splicing factor 3A ... 119 2e-27
cpv:cgd7_2160 Pre-mRNA splicing factor SF3a. 2xSWAP domain pro... 115 3e-26
dre:796388 sf3a1, MGC65786, MGC77239, si:dz150f13.2, wu:fc38e0... 108 4e-24
xla:444135 MGC80562 protein; K12825 splicing factor 3A subunit 1 107 8e-24
mmu:67465 Sf3a1, 1200014H24Rik, 5930416L09Rik, AI159724; splic... 102 5e-22
ath:AT1G14650 SWAP (Suppressor-of-White-APricot)/surp domain-c... 97.4 1e-20
ath:AT1G14640 SWAP (Suppressor-of-White-APricot)/surp domain-c... 92.4 4e-19
cel:W07E6.4 prp-21; yeast PRP (splicing factor) related family... 88.2 7e-18
pfa:PF14_0713 conserved Plasmodium protein, unknown function; ... 74.7 9e-14
hsa:10291 SF3A1, PRP21, PRPF21, SAP114, SF3A120; splicing fact... 66.6 2e-11
ath:AT5G06520 SWAP (Suppressor-of-White-APricot)/surp domain-c... 52.0 5e-07
ath:AT4G16200 SWAP (Suppressor-of-White-APricot)/surp domain-c... 47.4 1e-05
ath:AT3G49130 RNA binding 41.6 8e-04
ath:AT5G55100 SWAP (Suppressor-of-White-APricot)/surp domain-c... 38.1 0.008
mmu:27967 Cherp, 5730408I11Rik, D8Wsu96e, DAN16, Scaf6; calciu... 37.4 0.013
hsa:10523 CHERP, DAN16, SCAF6, SRA1; calcium homeostasis endop... 37.4 0.014
hsa:6433 SFSWAP, MGC167082, SFRS8, SWAP; splicing factor, supp... 35.8 0.036
ath:AT2G43960 SWAP (Suppressor-of-White-APricot)/surp domain-c... 35.8 0.036
ath:AT5G12280 RNA binding 34.7 0.081
mmu:231769 Sfswap, 1190005N23Rik, 6330437E22Rik, AI197402, AW2... 33.9 0.15
dre:394143 sfswap, MGC56184, sfrs8, srsf8, zgc:56184; splicing... 33.9 0.16
pfa:PF14_0028 pre-mRNA splicing factor, putative; K12842 U2-as... 33.9 0.17
dre:337851 cherp, MGC55518, ik:tdsubc_2h12, scaf6, wu:fc83d01,... 33.5 0.21
dre:557667 trio; si:dkey-158b13.2 (EC:2.7.11.1); K08810 triple... 33.1 0.25
dre:368847 trio, si:dz230d15.1; triple functional domain (PTPR... 33.1 0.25
hsa:7204 TRIO, ARHGEF23, FLJ42780, tgat; triple functional dom... 33.1 0.28
mmu:223435 Trio, 6720464I07Rik, AA408740, Solo; triple functio... 33.1 0.29
xla:447338 cherp, MGC83231; calcium homeostasis endoplasmic re... 32.3 0.40
ath:AT1G18890 ATCDPK1 (CALCIUM-DEPENDENT PROTEIN KINASE 1); ca... 31.6 0.67
xla:380137 cherp, MGC53695; calcium homeostasis endoplasmic re... 31.6 0.73
cel:F44F1.6 hypothetical protein 31.6 0.77
hsa:254225 RNF169, KIAA1991; ring finger protein 169 30.8
tgo:TGME49_070590 UBA/TS-N domain-containing protein (EC:3.2.1... 30.0 2.1
dre:100037380 zgc:163098; K12842 U2-associated protein SR140 30.0 2.3
mmu:52163 Camk1, AI505105, CaMKIalpha, D6Ertd263e; calcium/cal... 28.9 4.6
ath:AT4G31200 SWAP (Suppressor-of-White-APricot)/surp domain-c... 28.9 4.7
hsa:8536 CAMK1, CAMKI, MGC120317, MGC120318; calcium/calmoduli... 28.9 4.8
> tgo:TGME49_046500 surp module domain-containing protein ; K12825
splicing factor 3A subunit 1
Length=683
Score = 173 bits (438), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 84/138 (60%), Positives = 114/138 (82%), Gaps = 4/138 (2%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGV 60
NPY+A+Y+LKVREF TGE AP PQVPQAI DM++K++++++K++++LMLTQ E
Sbjct 104 NPYNAYYQLKVREFQTGEEAPRPQVPQAILDMRKKEEEERKKKERVLMLTQYGEE----A 159
Query 61 KEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKP 120
K+K+EPP P+ + + P++A +D+DII+ AQFVARNGQ+FL+GLAQRE+QNPQF FLKP
Sbjct 160 KKKIEPPAPDVYSVTQPYIALIDVDIIQTTAQFVARNGQRFLSGLAQRERQNPQFEFLKP 219
Query 121 SHHLFSYFASLVDAYTKC 138
+H LF YF +LVDAYTKC
Sbjct 220 THALFQYFTNLVDAYTKC 237
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 32/49 (65%), Gaps = 2/49 (4%)
Query 81 PVDL-DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
P DL II AQFVA+NG +F + +REQ N +FAFL P++ +Y+
Sbjct 63 PPDLRGIIEKTAQFVAKNGVEFEQRV-RREQNNQRFAFLFPNNPYNAYY 110
> bbo:BBOV_III005810 17.m07515; Surp module domain containing
protein; K12825 splicing factor 3A subunit 1
Length=482
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 64/139 (46%), Positives = 88/139 (63%), Gaps = 4/139 (2%)
Query 1 NPYHAFYKLKVREFTTG-EAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDG 59
N YHA+YKLK+ E G + P +PQAI D ++K + + Q +++LL LT S D
Sbjct 59 NAYHAYYKLKLTELRNGIDVEIKPSIPQAILDRRKKLELKNQFKERLLALTDFGSSSAD- 117
Query 60 VKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLK 119
+L P + F P+VA +D+DII+ A FVARNGQ+FL L +RE+ NPQF FL
Sbjct 118 --HELGEPETDTFSFTQPFVASMDMDIIKATACFVARNGQRFLVELTKREKNNPQFDFLN 175
Query 120 PSHHLFSYFASLVDAYTKC 138
PSH LF +F +L ++YTKC
Sbjct 176 PSHCLFGFFTNLTESYTKC 194
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 7/49 (14%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQN------PQFAFLKPSHHLFSYF 128
II AQFV++NG++F L + EQ + +FAFL P + +Y+
Sbjct 18 IIDKTAQFVSKNGEQFEQRL-RAEQSDGAAGAGAKFAFLSPDNAYHAYY 65
> tpv:TP02_0389 hypothetical protein; K12825 splicing factor 3A
subunit 1
Length=443
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 56/139 (40%), Positives = 88/139 (63%), Gaps = 6/139 (4%)
Query 1 NPYHAFYKLKVREFTTGEAAP-TPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDG 59
N YH +YKLK+ E + A P +PQAI D ++K + + + +++LL L+ S
Sbjct 58 NAYHLYYKLKLSELQGNKVADLNPSIPQAILDKREKLELKTKHKERLLALSDFRHS---- 113
Query 60 VKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLK 119
E ++ P + F P+++ +D ++I+ A FVARNG KFL L +RE+ NPQ+ FL
Sbjct 114 -SESVQKPEDDLFSFTMPFISALDYEVIKNTALFVARNGHKFLVDLNKREKNNPQYDFLN 172
Query 120 PSHHLFSYFASLVDAYTKC 138
PSH+LF++F++L D+YTKC
Sbjct 173 PSHYLFTFFSNLTDSYTKC 191
> cpv:cgd7_2160 Pre-mRNA splicing factor SF3a. 2xSWAP domain protein
; K12825 splicing factor 3A subunit 1
Length=462
Score = 115 bits (289), Expect = 3e-26, Method: Composition-based stats.
Identities = 56/142 (39%), Positives = 85/142 (59%), Gaps = 4/142 (2%)
Query 1 NPYHAFYKLKVREFTTGEAAPT--PQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKD 58
NP+H +YK ++ +F G + P +P+AI DM ++++Q ++++LMLT
Sbjct 54 NPFHLYYKKRIEDFKNGVSIDNSGPTIPRAILDMNSRKEKQIIAEKEVLMLTSFSGGFGF 113
Query 59 GVKEKLEPPPP--NQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFA 116
+EP P +Q+ + HP ++ D +I+ A ++ARNGQ FL+ L RE NPQF
Sbjct 114 MGGAVMEPEEPRKDQYTISHPIISIKDESVIKITAMYLARNGQSFLSDLTARESNNPQFD 173
Query 117 FLKPSHHLFSYFASLVDAYTKC 138
FLKP H LF YFA LV+AY+ C
Sbjct 174 FLKPGHALFGYFADLVEAYSLC 195
> dre:796388 sf3a1, MGC65786, MGC77239, si:dz150f13.2, wu:fc38e01,
wu:fc50a11, wu:fj37c05, zgc:65786; splicing factor 3a,
subunit 1; K12825 splicing factor 3A subunit 1
Length=780
Score = 108 bits (271), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 60/139 (43%), Positives = 86/139 (61%), Gaps = 14/139 (10%)
Query 1 NPYHAFYKLKVREFTTGEA-APTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDG 59
+PYHA+Y+ KV EF G+A P+ VP+ MQ QQ ++ Q Q++ T + +
Sbjct 79 DPYHAYYRHKVNEFKEGKAQEPSAAVPKV---MQHPQQLPQKVQAQVIHETVVPK----- 130
Query 60 VKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLK 119
EPPP +F+ P ++ DLD+++ AQFVARNG++FL L Q+EQ+N QF FL+
Sbjct 131 -----EPPPEFEFIADPPSISAFDLDVVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLR 185
Query 120 PSHHLFSYFASLVDAYTKC 138
P H LF+YF LV+ YTK
Sbjct 186 PQHSLFNYFTKLVEQYTKV 204
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
+I+ A FVARNG +F A + Q E NP+F FL PS +Y+ V+ +
Sbjct 42 NIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPSDPYHAYYRHKVNEF 92
> xla:444135 MGC80562 protein; K12825 splicing factor 3A subunit
1
Length=802
Score = 107 bits (268), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 60/139 (43%), Positives = 87/139 (62%), Gaps = 11/139 (7%)
Query 1 NPYHAFYKLKVREFTTGEA-APTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDG 59
+PYHA+Y+ KV EF G+A P+ +P+ ++ Q QQ ++ Q Q++ T I +
Sbjct 83 DPYHAYYRHKVNEFKEGKAQEPSAAIPKVMQQQQSVQQLPQKLQAQVVQETIIPK----- 137
Query 60 VKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLK 119
EPPP +FV P ++ DLD+++ AQFVARNG++FL L Q+EQ+N QF FL+
Sbjct 138 -----EPPPEYEFVADPPSISAFDLDVVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLR 192
Query 120 PSHHLFSYFASLVDAYTKC 138
P H LF+YF LV+ YTK
Sbjct 193 PQHSLFNYFTKLVEQYTKI 211
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
+I+ A FVARNG +F A + Q E NP+F FL P+ +Y+ V+ +
Sbjct 46 NIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAYYRHKVNEF 96
> mmu:67465 Sf3a1, 1200014H24Rik, 5930416L09Rik, AI159724; splicing
factor 3a, subunit 1; K12825 splicing factor 3A subunit
1
Length=791
Score = 102 bits (253), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 60/141 (42%), Positives = 88/141 (62%), Gaps = 13/141 (9%)
Query 1 NPYHAFYKLKVREFTTGEA-APTPQVPQAIRDMQQKQQQQKQK--QQQLLMLTQIDESEK 57
+PYHA+Y+ KV EF G+A P+ +P+ ++ QQ QQQ + Q Q++ T + +
Sbjct 88 DPYHAYYRHKVSEFKEGKAQEPSAAIPKVMQQQQQATQQQLPQKVQAQVIQETIVPK--- 144
Query 58 DGVKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAF 117
EPPP +F+ P ++ DLD+++ AQFVARNG++FL L Q+EQ+N QF F
Sbjct 145 -------EPPPEFEFIADPPSISAFDLDVVKLTAQFVARNGRQFLTQLMQKEQRNYQFDF 197
Query 118 LKPSHHLFSYFASLVDAYTKC 138
L+P H LF+YF LV+ YTK
Sbjct 198 LRPQHSLFNYFTKLVEQYTKI 218
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 65 EPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHL 124
E P P++ V+ + P +I+ A FVARNG +F A + Q E NP+F FL P+
Sbjct 31 EDPTPSKPVVGIIYPPPEVRNIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPY 90
Query 125 FSYFASLVDAY 135
+Y+ V +
Sbjct 91 HAYYRHKVSEF 101
> ath:AT1G14650 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein / ubiquitin family protein; K12825 splicing
factor 3A subunit 1
Length=785
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 53/150 (35%), Positives = 79/150 (52%), Gaps = 23/150 (15%)
Query 1 NPYHAFYKLKVREFTT-----------GEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLML 49
+PYHAFY+ K+ E+ + PQ+ D + Q Q Q +
Sbjct 107 DPYHAFYQHKLTEYRAQNKDGAQGTDDSDGTTDPQLDTGAADESEAGDTQPDLQAQFRIP 166
Query 50 TQIDESEKDGVKEKLEPPPPNQFVLQHP-WVAPVDLDIIRCCAQFVARNGQKFLAGLAQR 108
++ LE P P ++ ++ P + +LDII+ AQFVARNG+ FL GL+ R
Sbjct 167 SK-----------PLEAPEPEKYTVRLPEGITGEELDIIKLTAQFVARNGKSFLTGLSNR 215
Query 109 EQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
E NPQF F+KP+H +F++F SLVDAY++
Sbjct 216 ENNNPQFHFMKPTHSMFTFFTSLVDAYSEV 245
Score = 32.7 bits (73), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
I+ AQFV++NG +F + ++N +F FLK S +++ + Y
Sbjct 71 IVEKTAQFVSKNGLEFEKRIIVSNEKNAKFNFLKSSDPYHAFYQHKLTEY 120
> ath:AT1G14640 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=735
Score = 92.4 bits (228), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 55/147 (37%), Positives = 78/147 (53%), Gaps = 25/147 (17%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKD-- 58
NPYH FY+ KV E++ IRD Q + +L DES+
Sbjct 106 NPYHGFYRYKVTEYSC-----------HIRDGAQGTDVDDTEDPKL-----DDESDAKPD 149
Query 59 ------GVKEKLEPPPPNQFVLQHP-WVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQ 111
++ LE P P ++ ++ P + +LDII+ AQFVARNGQ FL L +RE
Sbjct 150 LQAQFRAPRKILEAPEPEKYTVRLPEGIMEAELDIIKHTAQFVARNGQSFLRELMRREVN 209
Query 112 NPQFAFLKPSHHLFSYFASLVDAYTKC 138
N QF F+KP+H +F++F SLVDAY++
Sbjct 210 NSQFQFMKPTHSMFTFFTSLVDAYSEV 236
Score = 33.5 bits (75), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLK---PSHHLFSY 127
I+ AQFV++NG F + + N F+FLK P H + Y
Sbjct 70 IVETTAQFVSQNGLAFGNKVKTEKANNANFSFLKSDNPYHGFYRY 114
> cel:W07E6.4 prp-21; yeast PRP (splicing factor) related family
member (prp-21); K12825 splicing factor 3A subunit 1
Length=655
Score = 88.2 bits (217), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 52/138 (37%), Positives = 70/138 (50%), Gaps = 24/138 (17%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGV 60
+PYHA+YK V +F+ G P+VPQA+++ +K +
Sbjct 73 DPYHAYYKKMVYDFSEGRVE-APKVPQAVKEHVKKAEFVPSA------------------ 113
Query 61 KEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKP 120
PPP +F + DLD+IR A FVARNG++FL L RE +N QF FLKP
Sbjct 114 -----PPPAYEFSADPSTINAYDLDLIRLVALFVARNGRQFLTQLMTREARNYQFDFLKP 168
Query 121 SHHLFSYFASLVDAYTKC 138
+H F+YF LVD Y K
Sbjct 169 AHCNFTYFTKLVDQYQKV 186
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 40/85 (47%), Gaps = 2/85 (2%)
Query 48 MLTQIDESEKDGVKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAGLAQ 107
M + E+D + EP + ++ + P I+ A+F A+NG F + +
Sbjct 1 MTAVVSNREEDSMNN--EPSLSGRAIIGLIYPPPDIRTIVDKTARFAAKNGVDFENKIRE 58
Query 108 REQQNPQFAFLKPSHHLFSYFASLV 132
+E +NP+F FL + +Y+ +V
Sbjct 59 KEAKNPKFNFLSITDPYHAYYKKMV 83
> pfa:PF14_0713 conserved Plasmodium protein, unknown function;
K12825 splicing factor 3A subunit 1
Length=701
Score = 74.7 bits (182), Expect = 9e-14, Method: Composition-based stats.
Identities = 46/159 (28%), Positives = 80/159 (50%), Gaps = 21/159 (13%)
Query 1 NPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQ--LLMLTQIDESEKD 58
+PY +Y+ K+ +P I+++++++ K + L + I E EK+
Sbjct 78 HPYFYYYQYKLHGLFLDIQEKDELIPHVIKEIKKREDANKYTNNEHILKICDFIKEDEKE 137
Query 59 GVK------------EKLEPPPPNQFVLQH-------PWVAPVDLDIIRCCAQFVARNGQ 99
K +KLE + +Q P++ +D+D+I+ A FVARNG+
Sbjct 138 ERKNIIYSLDDMKKIDKLENNNNLKIQIQEDIYSLISPFITSLDIDLIKTTALFVARNGK 197
Query 100 KFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
FL GL +RE+ N Q+ FL+ ++ F+YF+ L+D Y KC
Sbjct 198 SFLNGLIEREKNNSQYDFLRANNLYFNYFSKLIDIYLKC 236
Score = 32.3 bits (72), Expect = 0.44, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 25/43 (58%), Gaps = 2/43 (4%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
+I A FV +NG+ F + + +++ QF F+ PSH F Y+
Sbjct 44 VIDKTATFVKKNGKNFEQKIYREKEK--QFGFISPSHPYFYYY 84
> hsa:10291 SF3A1, PRP21, PRPF21, SAP114, SF3A120; splicing factor
3a, subunit 1, 120kDa; K12825 splicing factor 3A subunit
1
Length=728
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 30/48 (62%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 91 AQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
AQFVARNG++FL L Q+EQ+N QF FL+P H LF+YF LV+ YTK
Sbjct 106 AQFVARNGRQFLTQLMQKEQRNYQFDFLRPQHSLFNYFTKLVEQYTKI 153
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 1 NPYHAFYKLKVREFTTGEA 19
+PYHA+Y+ KV EF G+A
Sbjct 88 DPYHAYYRHKVSEFKEGKA 106
> ath:AT5G06520 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=679
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHH-LFSYFASLVDAYTKC 138
+L II+ AQF+AR G F+ GL +R NPQF FL+ +++ FS++ LV AY++
Sbjct 487 ELGIIKLTAQFMARYGMNFVQGLRKRVVGNPQFKFLESTNNSRFSFYNGLVIAYSRV 543
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
+LD I+ AQFVA G F L +R +P+F F K + S++ LVD Y++
Sbjct 218 ELDTIKLTAQFVAVYGTLFRTELMKRVFISPKFDFFKSTDSKCSFYLRLVDGYSRV 273
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAY 135
+L++I+ AQFVA G+ F L R ++P F FLKP+ S++ ++ Y
Sbjct 89 ELELIKLTAQFVAVYGKYFQRELTTRVVESPLFEFLKPTDSRNSFYTRIILGY 141
> ath:AT4G16200 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=288
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
+LD ++ AQFVA G F L +R PQF F+K + + FS++ V AY++
Sbjct 200 ELDTVKVTAQFVAWYGDDFRGFLMERVMTEPQFEFMKATDYRFSFYNEFVVAYSQV 255
> ath:AT3G49130 RNA binding
Length=307
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 45/94 (47%), Gaps = 3/94 (3%)
Query 45 QLLMLTQIDESEKDGVKEKLEPPPPNQFVLQHPWVAPVDLDIIRCCAQFVARNGQKFLAG 104
++++ ++ +++ D V L+ P +Q + + +L +++ AQ R G F G
Sbjct 24 SVMIMNRVAQTQPDMV---LQLPLGSQLTIPAKGITLKELGVMKLTAQSSVRYGFDFWCG 80
Query 105 LAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
L +R NP F FL PS ++ AY++
Sbjct 81 LWKRVYMNPLFQFLNPSDSRSDFYNGFTAAYSRV 114
Score = 31.2 bits (69), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 105 LAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
L ++ PQF F++P++ +F + +VDAY++
Sbjct 226 LMKKVVMKPQFKFMEPTNSVFGLYNVVVDAYSRV 259
> ath:AT5G55100 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=844
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 20/48 (41%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVD 133
II + FV+++G + L ++ NP F FL P HHL YF LVD
Sbjct 153 IITRTSSFVSKHGGQSEIVLRVKQGDNPTFGFLMPDHHLHLYFRFLVD 200
> mmu:27967 Cherp, 5730408I11Rik, D8Wsu96e, DAN16, Scaf6; calcium
homeostasis endoplasmic reticulum protein; K12841 calcium
homeostasis endoplasmic reticulum protein
Length=938
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 9/54 (16%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
++I AQFVARNG +F ++++ NP+F+FL F +Y KC
Sbjct 14 NVIDKLAQFVARNGPEFEKMTMEKQKDNPKFSFL---------FGGEFYSYYKC 58
> hsa:10523 CHERP, DAN16, SCAF6, SRA1; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic
reticulum protein
Length=916
Score = 37.4 bits (85), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 9/54 (16%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
++I AQFVARNG +F ++++ NP+F+FL F +Y KC
Sbjct 14 NVIDKLAQFVARNGPEFEKMTMEKQKDNPKFSFL---------FGGEFYSYYKC 58
> hsa:6433 SFSWAP, MGC167082, SFRS8, SWAP; splicing factor, suppressor
of white-apricot homolog (Drosophila)
Length=951
Score = 35.8 bits (81), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 31/67 (46%), Gaps = 15/67 (22%)
Query 77 PWVAPVDLD---------------IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPS 121
P+VAP+ L II A FV R G +F L ++ +N QF FL+
Sbjct 187 PFVAPLGLSVPSDVELPPTAKMHAIIERTASFVCRQGAQFEIMLKAKQARNSQFDFLRFD 246
Query 122 HHLFSYF 128
H+L Y+
Sbjct 247 HYLNPYY 253
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 2/43 (4%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
+I A++VARNG KF + R + + +F FL+P H +Y+
Sbjct 459 VIDKLAEYVARNGLKFETSV--RAKNDQRFEFLQPWHQYNAYY 499
> ath:AT2G43960 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=442
Score = 35.8 bits (81), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 26/54 (48%), Gaps = 0/54 (0%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYT 136
+L II+ AQFV R GQ F L R + +F FL +F+ L Y+
Sbjct 125 ELGIIKFTAQFVLRYGQYFRLALRDRVSTDTEFKFLDKGDSRAHFFSLLFLGYS 178
> ath:AT5G12280 RNA binding
Length=181
Score = 34.7 bits (78), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 92 QFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASL 131
+ V R G++F L + PQF FLKP+ F YF L
Sbjct 14 KIVGRYGEEFWLDLIKEVDNKPQFEFLKPADSKFDYFNRL 53
> mmu:231769 Sfswap, 1190005N23Rik, 6330437E22Rik, AI197402, AW212079,
SWAP, Sfrs8, Srsf8; splicing factor, suppressor of
white-apricot homolog (Drosophila)
Length=945
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 15/67 (22%)
Query 77 PWVAPVDLD---------------IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPS 121
P++AP+ L II A FV + G +F L ++ +N QF FL+
Sbjct 187 PFIAPLGLSVPSDVELPPTAKMHAIIERTANFVCKQGAQFEIMLKAKQARNSQFDFLRFD 246
Query 122 HHLFSYF 128
H+L Y+
Sbjct 247 HYLNPYY 253
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query 91 AQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
A++VARNG KF + R + + +F FL+P H +Y+
Sbjct 463 AEYVARNGLKFETSV--RAKNDQRFEFLQPWHQYNAYY 498
> dre:394143 sfswap, MGC56184, sfrs8, srsf8, zgc:56184; splicing
factor, suppressor of white-apricot homolog (Drosophila)
Length=958
Score = 33.9 bits (76), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query 91 AQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
A++VARNG KF + R + +P+F FL+ H SY+
Sbjct 483 AEYVARNGVKFETSV--RAKNDPRFDFLQSWHQYNSYY 518
Score = 33.5 bits (75), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDA 134
II A FV + G +F L ++ N QF FL+ H+L Y+ ++ A
Sbjct 199 IIERTANFVCKQGAQFEIVLKAKQAGNSQFDFLRFDHYLNPYYKHILRA 247
> pfa:PF14_0028 pre-mRNA splicing factor, putative; K12842 U2-associated
protein SR140
Length=655
Score = 33.9 bits (76), Expect = 0.17, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 86 IIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
II A++V G F + ++E+ NP F FL S LF Y+
Sbjct 294 IIDLLAKYVTEEGYSFEETIKEKEKDNPIFHFLFESSDLFYYY 336
> dre:337851 cherp, MGC55518, ik:tdsubc_2h12, scaf6, wu:fc83d01,
xx:tdsubc_2h12, zgc:55518; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic
reticulum protein
Length=909
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYF 128
++I AQFVARNG +F ++++ N +F+FL F Y+
Sbjct 14 NVIDKLAQFVARNGPEFEKMTMEKQKDNAKFSFLF-GGEFFGYY 56
> dre:557667 trio; si:dkey-158b13.2 (EC:2.7.11.1); K08810 triple
functional domain protein [EC:2.7.11.1]
Length=3087
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQR-EQQNPQFAFLKPSHHLFSYFASLVDAYTK 137
D+D+IR CA+ VA + Q+ + + R + N AF K S + S SL Y +
Sbjct 991 DMDMIRDCAENVASHWQQLMLKMEDRLKLVNASVAFYKTSEQVCSVLESLEQEYKR 1046
> dre:368847 trio, si:dz230d15.1; triple functional domain (PTPRF
interacting)
Length=1282
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQR-EQQNPQFAFLKPSHHLFSYFASLVDAYTK 137
D+D+IR CA+ VA + Q+ + + R + N AF K S + S SL Y +
Sbjct 1067 DMDMIRDCAEKVASHWQQLMLKMEDRLKLVNASVAFYKTSEQVCSVLESLEQEYKR 1122
> hsa:7204 TRIO, ARHGEF23, FLJ42780, tgat; triple functional domain
(PTPRF interacting) (EC:2.7.11.1); K08810 triple functional
domain protein [EC:2.7.11.1]
Length=3097
Score = 33.1 bits (74), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQR-EQQNPQFAFLKPSHHLFSYFASLVDAYTK 137
D+D+IR CA+ VA + Q+ + + R + N AF K S + S SL Y +
Sbjct 981 DMDMIRDCAEKVASHWQQLMLKMEDRLKLVNASVAFYKTSEQVCSVLESLEQEYKR 1036
> mmu:223435 Trio, 6720464I07Rik, AA408740, Solo; triple functional
domain (PTPRF interacting) (EC:2.7.11.1); K08810 triple
functional domain protein [EC:2.7.11.1]
Length=3103
Score = 33.1 bits (74), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 83 DLDIIRCCAQFVARNGQKFLAGLAQR-EQQNPQFAFLKPSHHLFSYFASLVDAYTK 137
D+D+IR CA+ VA + Q+ + + R + N AF K S + S SL Y +
Sbjct 981 DMDMIRDCAEKVASHWQQLMLKMEDRLKLVNASVAFYKTSEQVCSVLESLEQEYKR 1036
> xla:447338 cherp, MGC83231; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic reticulum
protein
Length=933
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 9/54 (16%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFLKPSHHLFSYFASLVDAYTKC 138
++I AQFVARNG +F ++++ N +F+FL F Y KC
Sbjct 14 NVIDKLAQFVARNGPEFEKMTMEKQKDNHKFSFL---------FGGEFYNYYKC 58
> ath:AT1G18890 ATCDPK1 (CALCIUM-DEPENDENT PROTEIN KINASE 1);
calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=545
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 51 QIDESEKDGVKEKLEPPPPN----QFVLQHPWV 79
QI ES K VK+ L+P P Q VL HPW+
Sbjct 289 QISESAKSLVKQMLDPDPTKRLTAQQVLAHPWI 321
> xla:380137 cherp, MGC53695; calcium homeostasis endoplasmic
reticulum protein
Length=924
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 85 DIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFL 118
++I AQFVARNG +F ++++ N +F+FL
Sbjct 14 NVIDKLAQFVARNGPEFEKMTMEKQKDNLKFSFL 47
> cel:F44F1.6 hypothetical protein
Length=493
Score = 31.6 bits (70), Expect = 0.77, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 2/53 (3%)
Query 7 YKLKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDG 59
Y + E+ AAP A+R ++KQ++ +++QQ ++ TQ E +KDG
Sbjct 356 YSIVTDEYVVLPAAPVKTF--AVRIQEEKQKKLDEEEQQKVLKTQELEKQKDG 406
> hsa:254225 RNF169, KIAA1991; ring finger protein 169
Length=708
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Query 9 LKVREFTTGEAAPTPQVPQAIRDMQQKQQQQKQKQQQLLMLTQIDESEKDGVKEK 63
LK T+GE P P +R+M+QK QQ+++ +Q L L ++ ++E+ V +
Sbjct 637 LKKLRQTSGEVGLAPTDP-VLREMEQKLQQEEEDRQLALQLQRMFDNERRTVSRR 690
> tgo:TGME49_070590 UBA/TS-N domain-containing protein (EC:3.2.1.11
2.7.11.1)
Length=5435
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 53 DESEKDGVKEKLEPPPPNQFVLQHPWVAPVDLDI 86
+ +E G ++KL PPP Q WVAP+ L +
Sbjct 1098 EPAESGGTQKKLRKPPPEQSCPSSLWVAPLVLPV 1131
> dre:100037380 zgc:163098; K12842 U2-associated protein SR140
Length=874
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 84 LDIIRCCAQFVARNGQKFLAGLAQREQQNPQFAFL---KPSHHLF 125
L +I +FV R G F A + RE+ NP F FL K H++
Sbjct 318 LGLIHRMIEFVVREGPMFEAMIMNREKNNPDFRFLFDNKSQEHVY 362
> mmu:52163 Camk1, AI505105, CaMKIalpha, D6Ertd263e; calcium/calmodulin-dependent
protein kinase I (EC:2.7.11.17); K08794
calcium/calmodulin-dependent protein kinase I [EC:2.7.11.17]
Length=374
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 4/34 (11%)
Query 51 QIDESEKDGVKEKLEPPPPNQFV----LQHPWVA 80
I +S KD ++ +E P +F LQHPW+A
Sbjct 244 DISDSAKDFIRHLMEKDPEKRFTCEQALQHPWIA 277
> ath:AT4G31200 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=650
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 87 IRCCAQFVARNGQKFLAGLAQREQQNPQFAFL 118
I ++ +NG +F A + R++ NP +AFL
Sbjct 130 IDKLVEYSVKNGPEFEAMMRDRQKDNPDYAFL 161
> hsa:8536 CAMK1, CAMKI, MGC120317, MGC120318; calcium/calmodulin-dependent
protein kinase I (EC:2.7.11.17); K08794 calcium/calmodulin-dependent
protein kinase I [EC:2.7.11.17]
Length=370
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 52 IDESEKDGVKEKLEPPPPNQFV----LQHPWVA 80
I +S KD ++ +E P +F LQHPW+A
Sbjct 245 ISDSAKDFIRHLMEKDPEKRFTCEQALQHPWIA 277
Lambda K H
0.320 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40