bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1253_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
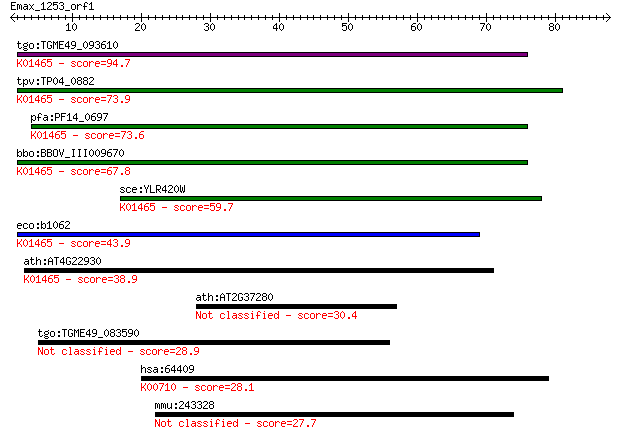
tgo:TGME49_093610 dihydroorotase protein, putative (EC:3.5.2.3... 94.7 7e-20
tpv:TP04_0882 dihydroorotase (EC:3.5.2.3); K01465 dihydroorota... 73.9 1e-13
pfa:PF14_0697 dihydroorotase, putative; K01465 dihydroorotase ... 73.6 2e-13
bbo:BBOV_III009670 17.m07839; dihydroorotase (EC:3.5.2.3); K01... 67.8 8e-12
sce:YLR420W URA4; Dihydroorotase, catalyzes the third enzymati... 59.7 2e-09
eco:b1062 pyrC, ECK1047, JW1049; dihydro-orotase (EC:3.5.2.3);... 43.9 1e-04
ath:AT4G22930 PYR4; PYR4 (PYRIMIDIN 4); dihydroorotase/ hydrol... 38.9 0.004
ath:AT2G37280 PDR5; PDR5 (PLEIOTROPIC DRUG RESISTANCE 5); ATPa... 30.4 1.4
tgo:TGME49_083590 hypothetical protein 28.9 4.5
hsa:64409 WBSCR17, DKFZp434I2216, DKFZp761D2324, GALNT16, GALN... 28.1 7.2
mmu:243328 Slc29a4, ENT4, MGC38048, mPMAT; solute carrier fami... 27.7 8.6
> tgo:TGME49_093610 dihydroorotase protein, putative (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=403
Score = 94.7 bits (234), Expect = 7e-20, Method: Composition-based stats.
Identities = 49/75 (65%), Positives = 54/75 (72%), Gaps = 1/75 (1%)
Query 2 GLTLHIHAESRKATALKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKG-KKNVA 60
GLTLH+H E L AEE F+P FEQIH RFP LKIV EH+STAAA++AVK NVA
Sbjct 150 GLTLHLHGEVPGVAPLDAEEAFIPFFEQIHSRFPSLKIVLEHVSTAAAIQAVKRMPANVA 209
Query 61 ATITAHHLRLTTGDV 75
ATIT HHL LT DV
Sbjct 210 ATITPHHLMLTVDDV 224
> tpv:TP04_0882 dihydroorotase (EC:3.5.2.3); K01465 dihydroorotase
[EC:3.5.2.3]
Length=295
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 52/79 (65%), Gaps = 0/79 (0%)
Query 2 GLTLHIHAESRKATALKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKGKKNVAA 61
G++LH+H E A L AE+ FLP E + ++F LK+V EH+++ +++ VK +N+AA
Sbjct 131 GVSLHLHGELPGAPPLTAEKEFLPIVENVCRKFSSLKVVLEHVTSKDSLQVVKRIQNLAA 190
Query 62 TITAHHLRLTTGDVWGQEE 80
TIT HH+RLT DV E
Sbjct 191 TITPHHMRLTVLDVLNTAE 209
> pfa:PF14_0697 dihydroorotase, putative; K01465 dihydroorotase
[EC:3.5.2.3]
Length=358
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 33/72 (45%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 4 TLHIHAESRKATALKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKGKKNVAATI 63
++HIH E L AEE +LP + +FPGL IV EHIS++ ++ +K +NVA +I
Sbjct 131 SIHIHCEEPNINPLYAEEKYLPHIHDLAIKFPGLNIVLEHISSSESINVIKEFRNVAGSI 190
Query 64 TAHHLRLTTGDV 75
T HHL LT DV
Sbjct 191 TPHHLYLTIDDV 202
> bbo:BBOV_III009670 17.m07839; dihydroorotase (EC:3.5.2.3); K01465
dihydroorotase [EC:3.5.2.3]
Length=351
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 2 GLTLHIHAESRKATALKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKGKKNVAA 61
GL+LHIH E + L++E F+ + FP LK+V EH+ST ++EAV N+AA
Sbjct 125 GLSLHIHGEQPGSNPLRSEAKFVQNIISVAMAFPKLKVVAEHVSTKESLEAVLRVPNLAA 184
Query 62 TITAHHLRLTTGDV 75
+IT HHL++ T DV
Sbjct 185 SITPHHLQIVTEDV 198
> sce:YLR420W URA4; Dihydroorotase, catalyzes the third enzymatic
step in the de novo biosynthesis of pyrimidines, converting
carbamoyl-L-aspartate into dihydroorotate (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=364
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 32/70 (45%), Positives = 44/70 (62%), Gaps = 9/70 (12%)
Query 17 LKAEETFLPAFEQIHQRFPGLKIVFEHISTAAAVEAVKG-KKN--------VAATITAHH 67
L AEE FLPA +++H FP LKI+ EH ++ +A++ ++ KN VAAT+TAHH
Sbjct 154 LNAEEAFLPALKKLHNDFPNLKIILEHCTSESAIKTIEDINKNVKKATDVKVAATLTAHH 213
Query 68 LRLTTGDVWG 77
L LT D G
Sbjct 214 LFLTIDDWAG 223
> eco:b1062 pyrC, ECK1047, JW1049; dihydro-orotase (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=348
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 28/71 (39%), Positives = 39/71 (54%), Gaps = 4/71 (5%)
Query 2 GLTLHIHAESRKATA--LKAEETFLPA-FEQIHQRFPGLKIVFEHISTAAAVEAVK-GKK 57
G+ L +H E A E F+ + E + QR LK+VFEHI+T A + V+ G +
Sbjct 134 GMPLLVHGEVTHADIDIFDREARFIESVMEPLRQRLTALKVVFEHITTKDAADYVRDGNE 193
Query 58 NVAATITAHHL 68
+AATIT HL
Sbjct 194 RLAATITPQHL 204
> ath:AT4G22930 PYR4; PYR4 (PYRIMIDIN 4); dihydroorotase/ hydrolase/
hydrolase, acting on carbon-nitrogen (but not peptide)
bonds, in cyclic amides (EC:3.5.2.3); K01465 dihydroorotase
[EC:3.5.2.3]
Length=377
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 38/73 (52%), Gaps = 5/73 (6%)
Query 3 LTLHIHAESRKAT--ALKAEETFLPAFEQ-IHQRFPGLKIVFEHISTAAAVEAVKGKK-- 57
+ L +H E + E+ F+ Q + QR P LK+V EHI+T AV V+ K
Sbjct 163 MPLLVHGEVTDPSIDVFDREKIFIETVLQPLIQRLPQLKVVMEHITTMDAVNFVESCKEG 222
Query 58 NVAATITAHHLRL 70
+V AT+T HL L
Sbjct 223 SVGATVTPQHLLL 235
> ath:AT2G37280 PDR5; PDR5 (PLEIOTROPIC DRUG RESISTANCE 5); ATPase,
coupled to transmembrane movement of substances
Length=1413
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 28 EQIHQRFPGLKIVFEHISTAAAVEAVKGK 56
E++ FP +++ +EH+ AA E V+GK
Sbjct 95 ERVGVEFPSIEVRYEHLGVEAACEVVEGK 123
> tgo:TGME49_083590 hypothetical protein
Length=380
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Query 5 LHIHAESRKATALKAEETFLPAFEQIH--QRFPGLKIVFEHISTAAAVEAVKG 55
LHI+ + + T K E LP +I+ P L + F+++ + A + VKG
Sbjct 142 LHIYLDQKIGTGKKEAEPLLPDLAEINAPDLMPTLVLNFDNVLASIAYDPVKG 194
> hsa:64409 WBSCR17, DKFZp434I2216, DKFZp761D2324, GALNT16, GALNT20,
GALNTL3; Williams-Beuren syndrome chromosome region 17
(EC:2.4.1.41); K00710 polypeptide N-acetylgalactosaminyltransferase
[EC:2.4.1.41]
Length=598
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 29/63 (46%), Gaps = 6/63 (9%)
Query 20 EETFLPAFEQIHQRFPGLKIVFEHISTAAAVEA-VKGKKNVAATITAH---HLRLTTGDV 75
EE +P E +H+R+PGL V + + A ++G K +T H+ T G
Sbjct 197 EELKVPLEEYVHKRYPGLVKVVRNQKREGLIRARIEGWKVATGQVTGFFDAHVEFTAG-- 254
Query 76 WGQ 78
W +
Sbjct 255 WAE 257
> mmu:243328 Slc29a4, ENT4, MGC38048, mPMAT; solute carrier family
29 (nucleoside transporters), member 4; K03323 solute carrier
family 29 (equilibrative nucleoside transporter), member
4
Length=528
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 22 TFLPAFEQIHQRFPGLKIVFEHISTAAAVE-AVKGKKNVAATITAHHLRLTTG 73
+F+ + +H ++PG IVF+ T V A NV H R+TTG
Sbjct 87 SFITDVDYLHHKYPGTSIVFDMSLTYILVALAAVLLNNVVVERLNLHTRITTG 139
Lambda K H
0.317 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026251472
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40