bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1248_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
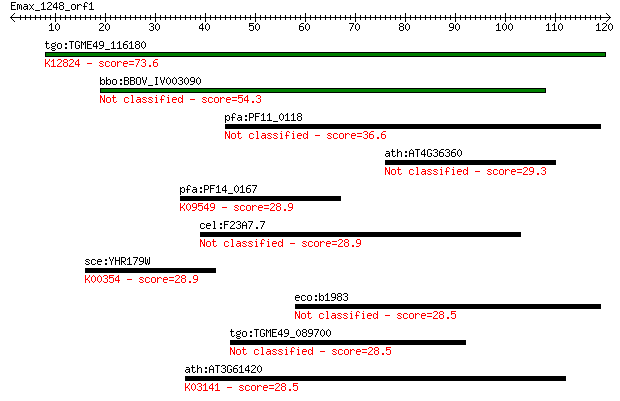
tgo:TGME49_116180 FF domain-containing protein ; K12824 transc... 73.6 2e-13
bbo:BBOV_IV003090 21.m02874; hypothetical protein 54.3 9e-08
pfa:PF11_0118 conserved Plasmodium protein 36.6 0.022
ath:AT4G36360 BGAL3; BGAL3 (beta-galactosidase 3); beta-galact... 29.3 2.9
pfa:PF14_0167 prefoldin subunit 2, putative; K09549 prefoldin ... 28.9 3.7
cel:F23A7.7 hypothetical protein 28.9 4.3
sce:YHR179W OYE2; Old Yellow Enzyme (EC:1.6.99.1); K00354 NADP... 28.9 4.6
eco:b1983 yeeN, ECK1978, JW1964; conserved protein, UPF0082 fa... 28.5 4.7
tgo:TGME49_089700 hypothetical protein 28.5 5.0
ath:AT3G61420 hypothetical protein; K03141 transcription initi... 28.5 5.4
> tgo:TGME49_116180 FF domain-containing protein ; K12824 transcription
elongation regulator 1
Length=772
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/112 (38%), Positives = 59/112 (52%), Gaps = 36/112 (32%)
Query 8 EKKKLKSLKADAATAFTNMLVERVKNPFLEAGEGNDLPTGLLKGDSRYTTNQLTETEKKE 67
E K+ + +K +AA AF NMLVERVKNPF
Sbjct 622 ETKRARLVKTEAAAAFMNMLVERVKNPFTS------------------------------ 651
Query 68 LYKAFVREFSTSRVSLFASRLSTLPTEDLNLSFEQVLEKLQTAKKLFHGISE 119
REF+T R+ LF ++L+TLP E L+ SF++VLE+LQT K+LF G+ +
Sbjct 652 ------REFTTGRLRLFQTKLNTLPCEKLSASFDEVLEELQTNKRLFDGLPQ 697
> bbo:BBOV_IV003090 21.m02874; hypothetical protein
Length=332
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 52/89 (58%), Gaps = 3/89 (3%)
Query 19 AATAFTNMLVERVKNPFLEAGEGNDLPTGLLKGDSRYTTNQLTETEKKELYKAFVREFST 78
A AF +ML ER++ PFL+ GE L LL+GD R + ++K LY FV EF
Sbjct 185 AHNAFMSMLHERIRMPFLD-GEIVPLDDDLLQGDPR--AEGCSAKDRKVLYDKFVSEFLE 241
Query 79 SRVSLFASRLSTLPTEDLNLSFEQVLEKL 107
+R++LF +LS + +E ++ S ++ E L
Sbjct 242 ARLALFDQKLSNIGSEQVSSSLDEKAELL 270
> pfa:PF11_0118 conserved Plasmodium protein
Length=594
Score = 36.6 bits (83), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 36/75 (48%), Gaps = 1/75 (1%)
Query 44 LPTGLLKGDSRYTTNQLTETEKKELYKAFVREFSTSRVSLFASRLSTLPTEDLNLSFEQV 103
LP +L D RY L + EK LYK F+ + S+ F LS L +N + +++
Sbjct 428 LPRDIL-NDERYKNITLNDNEKFVLYKEFINNYIDSKKISFHKLLSELSINCINNTLDEI 486
Query 104 LEKLQTAKKLFHGIS 118
+ + K+F I+
Sbjct 487 ILMIDKNNKMFKDIN 501
> ath:AT4G36360 BGAL3; BGAL3 (beta-galactosidase 3); beta-galactosidase/
catalytic/ cation binding / sugar binding
Length=856
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 76 FSTSRVSLFASRLSTLPTEDLNLSFEQVLEKLQT 109
F+T++V + S++ LPT+ N +E LE L +
Sbjct 418 FNTAKVGVQTSQMEMLPTDTKNFQWESYLEDLSS 451
> pfa:PF14_0167 prefoldin subunit 2, putative; K09549 prefoldin
subunit 2
Length=147
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 35 FLEAGEGNDLPTGLLKGDSRYTTNQLTETEKK 66
F+++ + +LP G++ G+ TN L + EKK
Sbjct 110 FIKSAKAGNLPGGIINGNPSEDTNNLNDKEKK 141
> cel:F23A7.7 hypothetical protein
Length=965
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 39 GEGNDLPTGLLKGDSRYTTNQLTETEKKELYKAFVREFSTSRVSLFASRLSTLPTEDLNL 98
G+ + P+G +K RY N L + LY A E SL ++ +S+L T L
Sbjct 852 GDLMNWPSGFIK--DRYDFNNLEQCLSGLLYSAIKLEDENVDTSLLSTAVSSLFTNHLKT 909
Query 99 SFEQ 102
FE
Sbjct 910 RFEH 913
> sce:YHR179W OYE2; Old Yellow Enzyme (EC:1.6.99.1); K00354 NADPH2
dehydrogenase [EC:1.6.99.1]
Length=400
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 16 KADAATAFTNMLVERVKNPFLEAGEG 41
KA AF +++ RV NPFL GEG
Sbjct 278 KAGKRLAFVHLVEPRVTNPFLTEGEG 303
> eco:b1983 yeeN, ECK1978, JW1964; conserved protein, UPF0082
family
Length=238
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 37/71 (52%), Gaps = 11/71 (15%)
Query 58 NQLTETEKKELYK-------AFVREFSTSRVSLFASRLSTLPTEDLNLSFEQVLEKLQ-- 108
N + TE +L+K A + EFST+ + + A L EDL + FE +++ L+
Sbjct 167 NIVIYTEPTDLHKGIAALKAAGITEFSTTELEMIAQSEVELSPEDLEI-FEGLVDALEDD 225
Query 109 -TAKKLFHGIS 118
+K++H ++
Sbjct 226 DDVQKVYHNVA 236
> tgo:TGME49_089700 hypothetical protein
Length=588
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query 45 PTGLLKGDSRYTTNQLTETEKKELYKAFVREFSTSRVSLFASRLSTL 91
P+ L GDSR T++L ++ LY+AF R S ++S STL
Sbjct 13 PSVRLHGDSREDTSRLERQQEAWLYRAFTRALLH---SFWSSSNSTL 56
> ath:AT3G61420 hypothetical protein; K03141 transcription initiation
factor TFIIH subunit 1
Length=579
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 16/76 (21%), Positives = 35/76 (46%), Gaps = 0/76 (0%)
Query 36 LEAGEGNDLPTGLLKGDSRYTTNQLTETEKKELYKAFVREFSTSRVSLFASRLSTLPTED 95
++A EG+D + G R TN + E + +L ++ +++ + + R + +ED
Sbjct 282 MDADEGDDYTHLMDHGIQRDGTNDIIEPQNDQLKRSLLQDLNRHAAVVLEGRCINVQSED 341
Query 96 LNLSFEQVLEKLQTAK 111
+ E + Q +K
Sbjct 342 TRIVAEALTRAKQVSK 357
Lambda K H
0.311 0.128 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40