bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1231_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
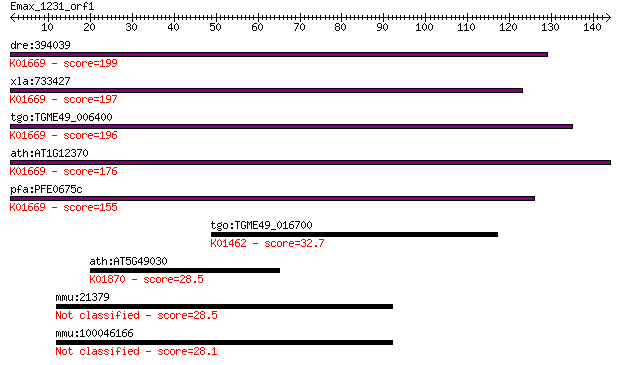
dre:394039 MGC66475, phr; zgc:66475; K01669 deoxyribodipyrimid... 199 2e-51
xla:733427 phr; CPD photolyase-like; K01669 deoxyribodipyrimid... 197 7e-51
tgo:TGME49_006400 DNA photolyase, putative (EC:4.1.99.3); K016... 196 2e-50
ath:AT1G12370 PHR1; PHR1 (PHOTOLYASE 1); DNA photolyase; K0166... 176 2e-44
pfa:PFE0675c deoxyribodipyrimidine photolyase (photoreactivati... 155 4e-38
tgo:TGME49_016700 peptide deformylase, putative (EC:3.5.1.88);... 32.7 0.36
ath:AT5G49030 OVA2; OVA2 (ovule abortion 2); ATP binding / ami... 28.5 7.4
mmu:21379 Tbrg4, 2310042P22Rik, AA120735, AA408001, AI527316, ... 28.5 7.4
mmu:100046166 protein TBRG4-like 28.1 9.4
> dre:394039 MGC66475, phr; zgc:66475; K01669 deoxyribodipyrimidine
photo-lyase [EC:4.1.99.3]
Length=516
Score = 199 bits (507), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 86/128 (67%), Positives = 107/128 (83%), Gaps = 0/128 (0%)
Query 1 HRGDKREYVYSLAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEWSPSAQEA 60
H D+R+Y+Y+ + E+A+THD LWNAAQ QL+ GKMHGFLRMYWAKKILEW+ S +EA
Sbjct 376 HAKDRRQYLYTKKELESAETHDQLWNAAQRQLLLEGKMHGFLRMYWAKKILEWTASPEEA 435
Query 61 LSAALHLNDKFSLDGTDPNGIVGCMWSVAGVHDQGWAERPVFGKIRYMNLSGCRRKFPVD 120
LS A++LND+ SLDG DPNG VGCMWS+ G+HDQGWAERP+FGKIRYMN +GC+RKF V+
Sbjct 436 LSIAIYLNDRLSLDGCDPNGYVGCMWSICGIHDQGWAERPIFGKIRYMNYAGCKRKFDVE 495
Query 121 EFVQKVQA 128
+F +K A
Sbjct 496 QFERKYAA 503
> xla:733427 phr; CPD photolyase-like; K01669 deoxyribodipyrimidine
photo-lyase [EC:4.1.99.3]
Length=557
Score = 197 bits (502), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 86/122 (70%), Positives = 101/122 (82%), Gaps = 0/122 (0%)
Query 1 HRGDKREYVYSLAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEWSPSAQEA 60
H DKR ++Y+L + EA KTHD LWNAAQLQ+VH GKMHGFLRMYWAKKILEW+ S +EA
Sbjct 425 HAKDKRTHLYTLEKLEAGKTHDPLWNAAQLQMVHEGKMHGFLRMYWAKKILEWTSSPEEA 484
Query 61 LSAALHLNDKFSLDGTDPNGIVGCMWSVAGVHDQGWAERPVFGKIRYMNLSGCRRKFPVD 120
L +L+LND++ LDG DPNG VGCMWS+ G+HDQGWAER VFGKIRYMN GC+RKF V
Sbjct 485 LHFSLYLNDRYELDGRDPNGYVGCMWSICGIHDQGWAERAVFGKIRYMNYQGCKRKFDVA 544
Query 121 EF 122
+F
Sbjct 545 QF 546
> tgo:TGME49_006400 DNA photolyase, putative (EC:4.1.99.3); K01669
deoxyribodipyrimidine photo-lyase [EC:4.1.99.3]
Length=631
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 90/134 (67%), Positives = 100/134 (74%), Gaps = 0/134 (0%)
Query 1 HRGDKREYVYSLAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEWSPSAQEA 60
H DKRE Y A EA KT+D LWNAAQLQLV GKMHG+LRMYWAKKILEW+ S QEA
Sbjct 398 HAKDKREPHYGFAALEAGKTYDELWNAAQLQLVRDGKMHGYLRMYWAKKILEWTKSPQEA 457
Query 61 LSAALHLNDKFSLDGTDPNGIVGCMWSVAGVHDQGWAERPVFGKIRYMNLSGCRRKFPVD 120
L A+ LNDK+ LDGTDPNG+ GCMWS+ GVHDQG+ ERPVFGKIRYMN GC RKF V
Sbjct 458 LKIAIALNDKYHLDGTDPNGVTGCMWSICGVHDQGFKERPVFGKIRYMNYPGCERKFDVK 517
Query 121 EFVQKVQASTRKLV 134
FV+K + V
Sbjct 518 AFVRKFPGAAENAV 531
> ath:AT1G12370 PHR1; PHR1 (PHOTOLYASE 1); DNA photolyase; K01669
deoxyribodipyrimidine photo-lyase [EC:4.1.99.3]
Length=490
Score = 176 bits (446), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 81/143 (56%), Positives = 105/143 (73%), Gaps = 7/143 (4%)
Query 1 HRGDKREYVYSLAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEWSPSAQEA 60
H DKRE++YSL Q E T D L++V+ GKMHGF+RMYWAKKILEW+ +EA
Sbjct 342 HASDKREHIYSLEQLEKGLTAD------PLEMVYQGKMHGFMRMYWAKKILEWTKGPEEA 395
Query 61 LSAALHLNDKFSLDGTDPNGIVGCMWSVAGVHDQGWAERPVFGKIRYMNLSGCRRKFPVD 120
LS +++LN+K+ +DG DP+G VGCMWS+ GVHDQGW ERPVFGKIRYMN +GC+RKF VD
Sbjct 396 LSISIYLNNKYEIDGRDPSGYVGCMWSICGVHDQGWKERPVFGKIRYMNYAGCKRKFNVD 455
Query 121 EFVQKVQASTRKLVKETLQAREQ 143
++ V+ S + K+ +A EQ
Sbjct 456 SYISYVK-SLVSVTKKKRKAEEQ 477
> pfa:PFE0675c deoxyribodipyrimidine photolyase (photoreactivating
enzyme, DNA photolyase), putative (EC:4.1.99.3); K01669
deoxyribodipyrimidine photo-lyase [EC:4.1.99.3]
Length=1113
Score = 155 bits (392), Expect = 4e-38, Method: Composition-based stats.
Identities = 67/125 (53%), Positives = 86/125 (68%), Gaps = 0/125 (0%)
Query 1 HRGDKREYVYSLAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEWSPSAQEA 60
H DKREY+Y F+ AKTHD LWN QLQL++ G +H FLRMYW KKIL WS +++ A
Sbjct 941 HDSDKREYLYDFDDFKNAKTHDDLWNCCQLQLINEGIIHEFLRMYWCKKILNWSGNSKTA 1000
Query 61 LSAALHLNDKFSLDGTDPNGIVGCMWSVAGVHDQGWAERPVFGKIRYMNLSGCRRKFPVD 120
L A+ LND FS+D P+G V M S+ G+HDQ W ER VFGKIR MN + C+R F ++
Sbjct 1001 LKCAMKLNDDFSIDAKSPHGYVSIMSSIMGIHDQSWNERTVFGKIRSMNYNSCKRLFDIN 1060
Query 121 EFVQK 125
++ K
Sbjct 1061 VYMSK 1065
> tgo:TGME49_016700 peptide deformylase, putative (EC:3.5.1.88);
K01462 peptide deformylase [EC:3.5.1.88]
Length=353
Score = 32.7 bits (73), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 40/71 (56%), Gaps = 6/71 (8%)
Query 49 KILEWSPSA--QEALSAALHLNDKF-SLDGTDPNGIVGCMWSVAGVHDQGWAERPVFGKI 105
+++ W+P+ +E+ + LN + SL G + + GC+ SV GV ERP+ ++
Sbjct 239 QMIVWNPTGDVRESSRERVFLNPRLLSLYGPLVSDVEGCL-SVPGVF--APVERPLHARV 295
Query 106 RYMNLSGCRRK 116
RY +L G +R+
Sbjct 296 RYTSLEGIQRE 306
> ath:AT5G49030 OVA2; OVA2 (ovule abortion 2); ATP binding / aminoacyl-tRNA
ligase/ catalytic/ isoleucine-tRNA ligase/ nucleotide
binding; K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=955
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 20 THDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEWSPSAQEALSAA 64
T D + AAQ+++ + G++ Y +K + WSPS++ AL+ A
Sbjct 254 TLDPEYEAAQIEVFGQMALKGYI--YRGRKPVHWSPSSRTALAEA 296
> mmu:21379 Tbrg4, 2310042P22Rik, AA120735, AA408001, AI527316,
Cpr2, R74877, Tb-12, mKIAA0948; transforming growth factor
beta regulated gene 4
Length=630
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 33/81 (40%), Gaps = 1/81 (1%)
Query 12 LAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEW-SPSAQEALSAALHLNDK 70
L E A D++W LQ VH ++H L + LE SP Q +H+N
Sbjct 400 LGSLEPALQVDLVWALCVLQHVHETELHTVLHPGLHARFLESKSPKDQSTFQKLVHINTT 459
Query 71 FSLDGTDPNGIVGCMWSVAGV 91
L+ + G +VA +
Sbjct 460 ALLEHPEYKGPFLPASAVAPI 480
> mmu:100046166 protein TBRG4-like
Length=535
Score = 28.1 bits (61), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 33/81 (40%), Gaps = 1/81 (1%)
Query 12 LAQFEAAKTHDVLWNAAQLQLVHVGKMHGFLRMYWAKKILEW-SPSAQEALSAALHLNDK 70
L E A D++W LQ VH ++H L + LE SP Q +H+N
Sbjct 305 LGSLEPALQVDLVWALCVLQHVHETELHTVLHPGLHARFLESKSPKDQSTFQKLVHINTT 364
Query 71 FSLDGTDPNGIVGCMWSVAGV 91
L+ + G +VA +
Sbjct 365 ALLEHPEYKGPFLPASAVAPI 385
Lambda K H
0.321 0.133 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40