bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1229_orf1
Length=202
Score E
Sequences producing significant alignments: (Bits) Value
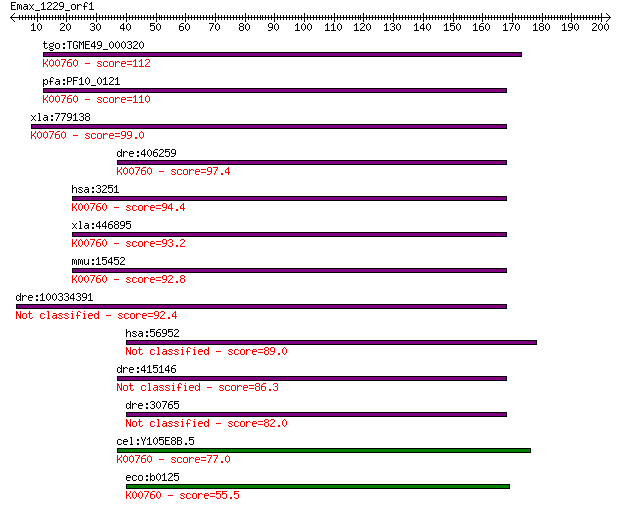
tgo:TGME49_000320 hypoxanthine-xanthine-guanine phosphoribosyl... 112 7e-25
pfa:PF10_0121 hypoxanthine phosphoribosyltransferase; K00760 h... 110 5e-24
xla:779138 hprt1, MGC82603, hgprt, hprt, prtfdc1; hypoxanthine... 99.0 1e-20
dre:406259 hprt1, id:ibd1344, id:ibd5108, wu:fc10g09, zgc:5622... 97.4 3e-20
hsa:3251 HPRT1, HGPRT, HPRT; hypoxanthine phosphoribosyltransf... 94.4 2e-19
xla:446895 prtfdc1, MGC80959; phosphoribosyl transferase domai... 93.2 5e-19
mmu:15452 Hprt, C81579, HPGRT, Hprt1, MGC103149; hypoxanthine ... 92.8 7e-19
dre:100334391 Hypoxanthine-guanine phosphoribosyltransferase-like 92.4 9e-19
hsa:56952 PRTFDC1, FLJ11888, HHGP; phosphoribosyl transferase ... 89.0 1e-17
dre:415146 hprt1l, zgc:86643; hypoxanthine phosphoribosyltrans... 86.3 7e-17
dre:30765 prtfdc1, HPRT, hprt1, hprt1l, zgc:55561, zgc:86771; ... 82.0 1e-15
cel:Y105E8B.5 hypothetical protein; K00760 hypoxanthine phosph... 77.0 4e-14
eco:b0125 hpt, ECK0124, JW5009; hypoxanthine phosphoribosyltra... 55.5 1e-07
> tgo:TGME49_000320 hypoxanthine-xanthine-guanine phosphoribosyl
transferase (EC:2.4.2.8); K00760 hypoxanthine phosphoribosyltransferase
[EC:2.4.2.8]
Length=279
Score = 112 bits (281), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 56/165 (33%), Positives = 100/165 (60%), Gaps = 5/165 (3%)
Query 12 PIHLQEKSSLYTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYYKEGELHLLC 71
P+++ + ++ Y A++ L+ P+I IL+P GL+ +R+EK+AYDI Y ELH++C
Sbjct 67 PMYIPD-NTFYNADDFLVPPHCKPYIDKILLPGGLVKDRVEKLAYDIHRTYFGEELHIIC 125
Query 72 LLKGARTFFG---DLTAAIYNHKSGCTKL-DIFHHFLQIRSYKNGEATEDVSFLGGDLST 127
+LKG+R FF D A I + + + F H+++++SY+N +T ++ L DLS
Sbjct 126 ILKGSRGFFNLLIDYLATIQKYSGRESSVPPFFEHYVRLKSYQNDNSTGQLTVLSDDLSI 185
Query 128 IKGKEVIIIDDVIETGKTITYIYKWLNKFNPRGVRTAALIQVEQD 172
+ K V+I++D+++TG T+T + L P+ +R A L++ D
Sbjct 186 FRDKHVLIVEDIVDTGFTLTEFGERLKAVGPKSMRIATLVEKRTD 230
> pfa:PF10_0121 hypoxanthine phosphoribosyltransferase; K00760
hypoxanthine phosphoribosyltransferase [EC:2.4.2.8]
Length=231
Score = 110 bits (274), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 57/160 (35%), Positives = 99/160 (61%), Gaps = 5/160 (3%)
Query 12 PIHLQEKSSLYTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYYKEGELHLLC 71
P+ +++ Y + ++ + + ++ +L+P+G+I NRIEK+AYDI Y E H+LC
Sbjct 16 PVFVKDDDG-YDLDSFMIPAHYKKYLTKVLVPNGVIKNRIEKLAYDIKKVYNNEEFHILC 74
Query 72 LLKGARTFFGDL---TAAIYNHKSGCTKLDIF-HHFLQIRSYKNGEATEDVSFLGGDLST 127
LLKG+R FF L + I+N+ + T +F H+++++SY N ++T + + DLS
Sbjct 75 LLKGSRGFFTALLKHLSRIHNYSAVETSKPLFGEHYVRVKSYCNDQSTGTLEIVSEDLSC 134
Query 128 IKGKEVIIIDDVIETGKTITYIYKWLNKFNPRGVRTAALI 167
+KGK V+I++D+I+TGKT+ ++L KF + V A L
Sbjct 135 LKGKHVLIVEDIIDTGKTLVKFCEYLKKFEIKTVAIACLF 174
> xla:779138 hprt1, MGC82603, hgprt, hprt, prtfdc1; hypoxanthine
phosphoribosyltransferase 1 (EC:2.4.2.8); K00760 hypoxanthine
phosphoribosyltransferase [EC:2.4.2.8]
Length=216
Score = 99.0 bits (245), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 55/162 (33%), Positives = 93/162 (57%), Gaps = 2/162 (1%)
Query 8 IGSVPIHLQEKSSLYTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYYKEGEL 67
+ S I +Q+ Y + + + ++ + IPHGLI++R E++A DI +
Sbjct 1 MASPCIVIQDDEQGYDLDLFCIPKHYAADLEKVYIPHGLIMDRTERLARDIMKDMGGHHI 60
Query 68 HLLCLLKGARTFFGDLTAAIYNHKSGCTK-LDIFHHFLQIRSYKNGEATEDVSFLGGD-L 125
LC+LKG FF DL I K + + F++++SY N ++T D+ +GGD L
Sbjct 61 VALCVLKGGYKFFADLLDYIKALNRNSDKSIPMTVDFIRLKSYCNDQSTGDIKVIGGDDL 120
Query 126 STIKGKEVIIIDDVIETGKTITYIYKWLNKFNPRGVRTAALI 167
ST+ GK V+I++D+I+TGKT+ + L K+NP+ V+ A+L+
Sbjct 121 STLTGKNVLIVEDIIDTGKTMKTLLAMLKKYNPKMVKVASLL 162
> dre:406259 hprt1, id:ibd1344, id:ibd5108, wu:fc10g09, zgc:56221,
zgc:86608; hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8);
K00760 hypoxanthine phosphoribosyltransferase [EC:2.4.2.8]
Length=218
Score = 97.4 bits (241), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 49/133 (36%), Positives = 87/133 (65%), Gaps = 2/133 (1%)
Query 37 IKGILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFGDLTAAIYN-HKSGCT 95
++ + IPHGLI++R E++A DI + LC+LKG FF DL I +++
Sbjct 32 LERVYIPHGLIMDRTERLARDIMKDMGGHHIVALCVLKGGYKFFADLLDYIKALNRNSDR 91
Query 96 KLDIFHHFLQIRSYKNGEATEDVSFLGGD-LSTIKGKEVIIIDDVIETGKTITYIYKWLN 154
+ + F++++SY+N ++T D+ +GGD LST+ GK V+I++D+I+TGKT+ + + L
Sbjct 92 SIPMTVDFIRLKSYQNDQSTGDIKVIGGDDLSTLTGKNVLIVEDIIDTGKTMKTLLELLK 151
Query 155 KFNPRGVRTAALI 167
++NP+ V+ A+L+
Sbjct 152 QYNPKMVKVASLL 164
> hsa:3251 HPRT1, HGPRT, HPRT; hypoxanthine phosphoribosyltransferase
1 (EC:2.4.2.8); K00760 hypoxanthine phosphoribosyltransferase
[EC:2.4.2.8]
Length=218
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 90/148 (60%), Gaps = 2/148 (1%)
Query 22 YTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFG 81
Y + + + + ++ + IPHGLI++R E++A D+ + LC+LKG FF
Sbjct 17 YDLDLFCIPNHYAEDLERVFIPHGLIMDRTERLARDVMKEMGGHHIVALCVLKGGYKFFA 76
Query 82 DLTAAIYN-HKSGCTKLDIFHHFLQIRSYKNGEATEDVSFLGGD-LSTIKGKEVIIIDDV 139
DL I +++ + + F++++SY N ++T D+ +GGD LST+ GK V+I++D+
Sbjct 77 DLLDYIKALNRNSDRSIPMTVDFIRLKSYCNDQSTGDIKVIGGDDLSTLTGKNVLIVEDI 136
Query 140 IETGKTITYIYKWLNKFNPRGVRTAALI 167
I+TGKT+ + + ++NP+ V+ A+L+
Sbjct 137 IDTGKTMQTLLSLVRQYNPKMVKVASLL 164
> xla:446895 prtfdc1, MGC80959; phosphoribosyl transferase domain
containing 1 (EC:2.4.2.8); K00760 hypoxanthine phosphoribosyltransferase
[EC:2.4.2.8]
Length=224
Score = 93.2 bits (230), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 45/148 (30%), Positives = 88/148 (59%), Gaps = 2/148 (1%)
Query 22 YTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFG 81
Y + L + + ++ + IPHG+I++R E++A+DI + + +LC+LKG F
Sbjct 23 YELDVFSLPNHYCEDLECVFIPHGVIVDRTERIAHDIMRDIGDNHITVLCVLKGGYRFCT 82
Query 82 DLTAAIYNHKSGCTK-LDIFHHFLQIRSYKNGEATEDVSFLGG-DLSTIKGKEVIIIDDV 139
DL I N + + + F++++ Y+N + +++ +GG DL+ + GK V+I++D+
Sbjct 83 DLVEHIKNLSRNSERFISMRVDFIRLKCYRNDQCMDEMQIIGGEDLAKLSGKNVLIVEDI 142
Query 140 IETGKTITYIYKWLNKFNPRGVRTAALI 167
I TG+T+T + L K+ P+ V+ A+L+
Sbjct 143 INTGRTMTALLNQLEKYKPKMVKVASLL 170
> mmu:15452 Hprt, C81579, HPGRT, Hprt1, MGC103149; hypoxanthine
guanine phosphoribosyl transferase (EC:2.4.2.8); K00760 hypoxanthine
phosphoribosyltransferase [EC:2.4.2.8]
Length=218
Score = 92.8 bits (229), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 47/148 (31%), Positives = 90/148 (60%), Gaps = 2/148 (1%)
Query 22 YTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFG 81
Y + + + + ++ + IPHGLI++R E++A D+ + LC+LKG FF
Sbjct 17 YDLDLFCIPNHYAEDLEKVFIPHGLIMDRTERLARDVMKEMGGHHIVALCVLKGGYKFFA 76
Query 82 DLTAAIYN-HKSGCTKLDIFHHFLQIRSYKNGEATEDVSFLGGD-LSTIKGKEVIIIDDV 139
DL I +++ + + F++++SY N ++T D+ +GGD LST+ GK V+I++D+
Sbjct 77 DLLDYIKALNRNSDRSIPMTVDFIRLKSYCNDQSTGDIKVIGGDDLSTLTGKNVLIVEDI 136
Query 140 IETGKTITYIYKWLNKFNPRGVRTAALI 167
I+TGKT+ + + +++P+ V+ A+L+
Sbjct 137 IDTGKTMQTLLSLVKQYSPKMVKVASLL 164
> dre:100334391 Hypoxanthine-guanine phosphoribosyltransferase-like
Length=221
Score = 92.4 bits (228), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 54/167 (32%), Positives = 98/167 (58%), Gaps = 5/167 (2%)
Query 3 EKKKKIGSVPIHLQEKSSLYTAEEILLSSSFLPFIKGILIPHGLIINRIEKMAYDICNYY 62
+++KK G V +++ + Y+ E + ++ + IPHG+I+NRIE +A DI
Sbjct 4 DQRKKPGVV---IKDGWTGYSLELFNYPEHYKGDLECVYIPHGVIMNRIECLARDILEDI 60
Query 63 KEGELHLLCLLKGARTFFGDLTAAIYNH-KSGCTKLDIFHHFLQIRSYKNGEATEDVSFL 121
++ +LC+LKG F DL +I +S ++L F++ +SY N ++TED+ +
Sbjct 61 GHHDMMVLCVLKGGYKFCSDLVESIKAQSRSTNSRLTTRVEFIRFKSYLNDQSTEDLHII 120
Query 122 G-GDLSTIKGKEVIIIDDVIETGKTITYIYKWLNKFNPRGVRTAALI 167
G DLS +KGK V+I++ +++TGKT+ + + + F P+ V+ A L+
Sbjct 121 GPDDLSMLKGKNVLIVEAIVDTGKTMRALLQHVETFQPKMVKVAGLL 167
> hsa:56952 PRTFDC1, FLJ11888, HHGP; phosphoribosyl transferase
domain containing 1 (EC:2.4.2.8)
Length=225
Score = 89.0 bits (219), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 85/143 (59%), Gaps = 5/143 (3%)
Query 40 ILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFGDLTAAIYNHKSGCTK-LD 98
+LIPHG+I++RIE++A DI ++ +LC+LKG F DL + N + +
Sbjct 42 VLIPHGIIVDRIERLAKDIMKDIGYSDIMVLCVLKGGYKFCADLVEHLKNISRNSDRFVS 101
Query 99 IFHHFLQIRSYKNGEATEDVSFLGG-DLSTIKGKEVIIIDDVIETGKTITYIYKWLNKFN 157
+ F++++SY+N ++ ++ +GG DLST+ GK V+I++DV+ TG+T+ + + K+
Sbjct 102 MKVDFIRLKSYRNDQSMGEMQIIGGDDLSTLAGKNVLIVEDVVGTGRTMKALLSNIEKYK 161
Query 158 PRGVRTAALI---QVEQDGVAPS 177
P ++ A+L+ DG P
Sbjct 162 PNMIKVASLLVKRTSRSDGFRPD 184
> dre:415146 hprt1l, zgc:86643; hypoxanthine phosphoribosyltransferase
1, like
Length=215
Score = 86.3 bits (212), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 80/135 (59%), Gaps = 6/135 (4%)
Query 37 IKGILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFGDLTAAIY---NHKSG 93
+ ++IP+GLI +R E++A DI + LC+LKG FF DL I H
Sbjct 29 LDSVIIPNGLIKDRTERLARDIVRDMGGHHIVALCVLKGGYKFFADLMDFIKTLNQHSDK 88
Query 94 CTKLDIFHHFLQIRSYKNGEATEDVSFLGGD-LSTIKGKEVIIIDDVIETGKTITYIYKW 152
L + F++++SY N ++T V +GGD LS + GK V+I++D++ETGKT+ + K
Sbjct 89 SVPLTV--DFIRLKSYCNDQSTNCVKVIGGDELSALAGKNVLIVEDIVETGKTMETLLKL 146
Query 153 LNKFNPRGVRTAALI 167
L++ +P+ V+ +L+
Sbjct 147 LHECHPKMVKVVSLL 161
> dre:30765 prtfdc1, HPRT, hprt1, hprt1l, zgc:55561, zgc:86771;
phosphoribosyl transferase domain containing 1
Length=225
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 80/133 (60%), Gaps = 8/133 (6%)
Query 40 ILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFGDLTAAIYNHKSGCTK--- 96
+ IPHG+I++RIE++A +I + + ++ +LC+LKG F DL I K C
Sbjct 42 VYIPHGVIMDRIERLARNIMDDLGDHDIVVLCVLKGGYQFCADLVDCI---KVLCCNSNK 98
Query 97 -LDIFHHFLQIRSYKNGEATEDVSFLGGD-LSTIKGKEVIIIDDVIETGKTITYIYKWLN 154
L + F++++SY N ++TED+ G + LS + GK V+I++ +++TGKT+ + +
Sbjct 99 TLPMRVDFIRLKSYLNDQSTEDLHIEGAENLSALSGKNVLIVEAIVDTGKTMKALLDHVE 158
Query 155 KFNPRGVRTAALI 167
F P+ ++ A L+
Sbjct 159 AFKPKMIKVAGLL 171
> cel:Y105E8B.5 hypothetical protein; K00760 hypoxanthine phosphoribosyltransferase
[EC:2.4.2.8]
Length=214
Score = 77.0 bits (188), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 45/151 (29%), Positives = 83/151 (54%), Gaps = 16/151 (10%)
Query 37 IKGILIPHGLIINRIEKMAYDICNYYKEGELHLLCLLKGARTFFGDLTAAIYNHKSGCTK 96
+ G++IP GL+ +R+ ++A DI + LLC+LKG+ FF L + N +S C +
Sbjct 29 LSGVVIPEGLVRDRVRRLAKDIHAEIGNKPIALLCVLKGSYKFFTALVEELTNARSSCPE 88
Query 97 LDIFHHFLQIRSYKNGEATEDVSFLG-GDLSTIKGKEVIIIDDVIETGKTITYIYKWLNK 155
+ F++++SY++ +T + +G +L +KGK V+++DD+ +TG+T+ K L+
Sbjct 89 -PMTVDFIRVKSYEDQMSTGQIQIMGLSNLDELKGKSVLVVDDISDTGRTLA---KLLST 144
Query 156 FNPRGV-----------RTAALIQVEQDGVA 175
+ GV R ++ V +D VA
Sbjct 145 LHETGVEKTWTALLLSKRVKRVVDVPEDFVA 175
> eco:b0125 hpt, ECK0124, JW5009; hypoxanthine phosphoribosyltransferase
(EC:2.4.2.8); K00760 hypoxanthine phosphoribosyltransferase
[EC:2.4.2.8]
Length=178
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 69/134 (51%), Gaps = 15/134 (11%)
Query 40 ILIPHGLIINRIEKMAYDICNYYKEG--ELHLLCLLKGARTFFGDLTAAIYNHKSGCTKL 97
++IP I RI ++ I YK+ ++ L+ LL+G+ F DL C ++
Sbjct 7 VMIPEAEIKARIAELGRQITERYKDSGSDMVLVGLLRGSFMFMADL----------CREV 56
Query 98 DIFHH--FLQIRSYKNGEAT-EDVSFLGGDLSTIKGKEVIIIDDVIETGKTITYIYKWLN 154
+ H F+ SY +G +T DV L I+GK+V+I++D+I++G T++ + + L+
Sbjct 57 QVSHEVDFMTASSYGSGMSTTRDVKILKDLDEDIRGKDVLIVEDIIDSGNTLSKVREILS 116
Query 155 KFNPRGVRTAALIQ 168
P+ + L+
Sbjct 117 LREPKSLAICTLLD 130
Lambda K H
0.314 0.131 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6124680020
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40