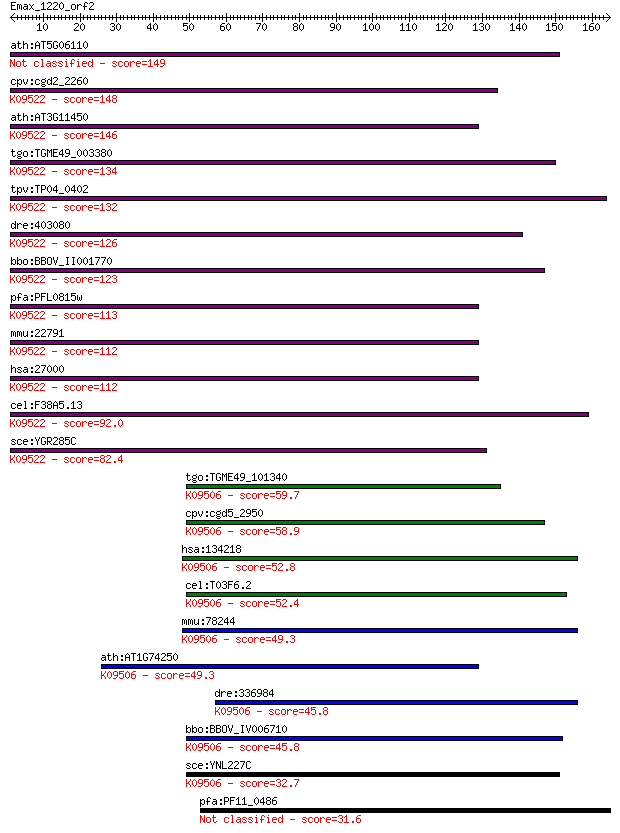
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1220_orf2
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
ath:AT5G06110 DNAJ heat shock N-terminal domain-containing pro... 149 3e-36
cpv:cgd2_2260 zuotin related factor-1 like protein with a DNAJ... 148 9e-36
ath:AT3G11450 DNAJ heat shock N-terminal domain-containing pro... 146 3e-35
tgo:TGME49_003380 DnaJ domain-containing protein ; K09522 DnaJ... 134 1e-31
tpv:TP04_0402 DNA-binding chaperone; K09522 DnaJ homolog subfa... 132 7e-31
dre:403080 dnajc2, zgc:85671, zrf1; DnaJ (Hsp40) homolog, subf... 126 3e-29
bbo:BBOV_II001770 18.m06137; myb-like DNA-binding/DnaJ domain ... 123 3e-28
pfa:PFL0815w DNA-binding chaperone, putative; K09522 DnaJ homo... 113 4e-25
mmu:22791 Dnajc2, AU020218, MIDA1, Zrf1, Zrf2; DnaJ (Hsp40) ho... 112 5e-25
hsa:27000 DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1; DnaJ (Hsp40) h... 112 5e-25
cel:F38A5.13 dnj-11; DNaJ domain (prokaryotic heat shock prote... 92.0 8e-19
sce:YGR285C ZUO1; Zuo1p; K09522 DnaJ homolog subfamily C member 2 82.4
tgo:TGME49_101340 DnaJ / zinc finger (C2H2 type) domain-contai... 59.7 4e-09
cpv:cgd5_2950 DNAj domain protein ; K09506 DnaJ homolog subfam... 58.9 7e-09
hsa:134218 DNAJC21, DNAJA5, GS3, JJJ1; DnaJ (Hsp40) homolog, s... 52.8 5e-07
cel:T03F6.2 dnj-17; DNaJ domain (prokaryotic heat shock protei... 52.4 7e-07
mmu:78244 Dnajc21, 4930461P20Rik, 9930116P15Rik; DnaJ (Hsp40) ... 49.3 5e-06
ath:AT1G74250 DNAJ heat shock N-terminal domain-containing pro... 49.3 5e-06
dre:336984 dnajc21, fa66c02, wu:fa66c02, zgc:63563; DnaJ (Hsp4... 45.8 6e-05
bbo:BBOV_IV006710 23.m06083; DnaJ domain containing protein; K... 45.8 7e-05
sce:YNL227C JJJ1; Co-chaperone that stimulates the ATPase acti... 32.7 0.53
pfa:PF11_0486 MAEBL, putative 31.6 1.4
> ath:AT5G06110 DNAJ heat shock N-terminal domain-containing protein
/ cell division protein-related
Length=663
Score = 149 bits (377), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 77/150 (51%), Positives = 109/150 (72%), Gaps = 4/150 (2%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
RR +DS FD+ VP+ DC +P+DF+K+FG AF+RNARWS N P+ LGD +TPL V+
Sbjct 175 RRIFDSTDEFDDKVPT--DC-APQDFFKVFGPAFKRNARWS-NSPLPDLGDENTPLKEVD 230
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
FY W+ F+SWR+F +EHD++QA R E+RWMERENAR +K K E ARI+ LV+
Sbjct 231 RFYSTWYTFKSWREFPEEEEHDIEQAESREEKRWMERENARKTQKARKEEYARIRTLVDN 290
Query 121 AHAADPRIKQRKEEERKKKEEEKAAQLRAR 150
A+ D RI++RK++E+ KK ++K A++ A+
Sbjct 291 AYKKDIRIQKRKDDEKAKKLQKKEAKVMAK 320
> cpv:cgd2_2260 zuotin related factor-1 like protein with a DNAJ
domain at the N-terminus and 2 SANT domains ; K09522 DnaJ
homolog subfamily C member 2
Length=677
Score = 148 bits (373), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 68/136 (50%), Positives = 95/136 (69%), Gaps = 4/136 (2%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKI---FGAAFQRNARWSVNRPVLTLGDCSTPLS 57
R YDSALPFD+S+PS ++ DFY+ F F+RN+RWS+ +PV +G+ +
Sbjct 177 RHAYDSALPFDDSIPSVYVSEN-NDFYEFKDFFSPIFRRNSRWSIVKPVPEIGNIDDNIE 235
Query 58 AVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKL 117
+E FYEFW F+S RDF +H+EH+L+ A CR E+RWMER+N +I+ KYI+NE +RI +L
Sbjct 236 VIESFYEFWRGFQSNRDFSIHEEHELNHAECREEKRWMERQNFKIRSKYIRNEISRINRL 295
Query 118 VETAHAADPRIKQRKE 133
V+ A+ DPRIKQ E
Sbjct 296 VDLAYKNDPRIKQHFE 311
> ath:AT3G11450 DNAJ heat shock N-terminal domain-containing protein
/ cell division protein-related; K09522 DnaJ homolog
subfamily C member 2
Length=663
Score = 146 bits (369), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 73/128 (57%), Positives = 94/128 (73%), Gaps = 3/128 (2%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
RR +DS FD+ VPS DC P+DF+K+FG AF+RNARWSVN+ + LGD +TPL V+
Sbjct 190 RRIFDSTDEFDDEVPS--DCL-PQDFFKVFGPAFKRNARWSVNQRIPDLGDENTPLKDVD 246
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
+FY FW+AF+SWR+F +EHDL+QA R ERRWME+ENA+ K K E ARI+ LV+
Sbjct 247 KFYNFWYAFKSWREFPDEEEHDLEQADSREERRWMEKENAKKTVKARKEEHARIRTLVDN 306
Query 121 AHAADPRI 128
A+ DPRI
Sbjct 307 AYRKDPRI 314
> tgo:TGME49_003380 DnaJ domain-containing protein ; K09522 DnaJ
homolog subfamily C member 2
Length=684
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 81/149 (54%), Positives = 98/149 (65%), Gaps = 30/149 (20%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
RRQYDSALP WS RPV +LGD TP+S V
Sbjct 224 RRQYDSALP------------------------------WSSRRPVPSLGDAQTPMSRVR 253
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
FY+FWF F+SWRDFGVHDE+DL++A CR ERRWMEREN +I+KK++K ERARIQKLVET
Sbjct 254 SFYDFWFDFQSWRDFGVHDEYDLNEAECREERRWMERENLKIRKKHVKAERARIQKLVET 313
Query 121 AHAADPRIKQRKEEERKKKEEEKAAQLRA 149
A++ DPR+ KE +KK+EEEKAA+ R
Sbjct 314 AYSVDPRVLMEKESAKKKREEEKAARQRV 342
> tpv:TP04_0402 DNA-binding chaperone; K09522 DnaJ homolog subfamily
C member 2
Length=674
Score = 132 bits (331), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 72/165 (43%), Positives = 107/165 (64%), Gaps = 3/165 (1%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
R +YDS LPFDES+P+ + + +DF+ +FG FQ N+RWS +PV LGD + V+
Sbjct 220 RLEYDSNLPFDESIPTHEEAKR-KDFFLLFGPIFQMNSRWSRVKPVPLLGDNESDDEYVD 278
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
+FYEFW FE+ R F H L+ A R E+RWMEREN +++KK IK E+ RIQKL++
Sbjct 279 DFYEFWRCFETLRTFSHAAPHLLEDAESREEKRWMERENLKVQKKLIKKEQLRIQKLIDL 338
Query 121 AHAADPRIKQRKEEERKKK--EEEKAAQLRAREEARRAAEDQKRR 163
+ DPRIK+R++ R +K ++++A + + E R E +K+R
Sbjct 339 SLQYDPRIKRRQDRIRHEKLVKQKQAQEAKLLELRRIELEKEKQR 383
> dre:403080 dnajc2, zgc:85671, zrf1; DnaJ (Hsp40) homolog, subfamily
C, member 2; K09522 DnaJ homolog subfamily C member
2
Length=618
Score = 126 bits (316), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 64/141 (45%), Positives = 95/141 (67%), Gaps = 3/141 (2%)
Query 1 RRQYDSALP-FDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
RR +DS P FD +VP+ A+ + E+F+++F F+RNARWSV + +LG + V
Sbjct 151 RRAFDSVDPTFDNAVPTKAEGK--ENFFEVFAPVFERNARWSVKKHFPSLGTMESSFEDV 208
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVE 119
+ FY FW+ F+SWR+F DE + ++A CR ERRW+E++N + + K E RI+ LV+
Sbjct 209 DNFYSFWYNFDSWREFSYLDEEEKEKAECRDERRWIEKQNRASRAQRKKEEMNRIRTLVD 268
Query 120 TAHAADPRIKQRKEEERKKKE 140
TA+ ADPRIK+ KEEE+ +KE
Sbjct 269 TAYNADPRIKKFKEEEKARKE 289
> bbo:BBOV_II001770 18.m06137; myb-like DNA-binding/DnaJ domain
containing protein; K09522 DnaJ homolog subfamily C member
2
Length=647
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 66/146 (45%), Positives = 93/146 (63%), Gaps = 0/146 (0%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
R +YD +LPFDE++P+ + + +DFY +F F+ NARWS +PV +LG ++ ++
Sbjct 220 RHEYDCSLPFDETIPTKEEAKLADDFYGLFAPVFELNARWSRTKPVPSLGAANSSDDDID 279
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
FY+FW FE+ R F H LD A R E+RWMEREN ++++K IK E RIQKLV+
Sbjct 280 FFYDFWRNFETTRTFSHAAPHLLDDAESREEKRWMERENLKVQRKLIKKELVRIQKLVDL 339
Query 121 AHAADPRIKQRKEEERKKKEEEKAAQ 146
A A DPR+K R E ++K E K Q
Sbjct 340 AQAFDPRLKARAERRLQEKVERKRLQ 365
> pfa:PFL0815w DNA-binding chaperone, putative; K09522 DnaJ homolog
subfamily C member 2
Length=939
Score = 113 bits (282), Expect = 4e-25, Method: Composition-based stats.
Identities = 63/128 (49%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
R+QYDS++PFDE +P+ + ++FY F+RNA+WS +PV +GD +T + V+
Sbjct 291 RKQYDSSIPFDERIPTLTQLEEAKNFYNFLRPVFKRNAKWSAIKPVPDIGDENTDIKEVK 350
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVET 120
FY+FW+ F +WRDF +E+D +QA CR ERRWMEREN +I+KK K E RI KLV+
Sbjct 351 YFYDFWYNFNNWRDFSYQNEYDYEQAECREERRWMERENKKIQKKASKTENLRIIKLVDL 410
Query 121 AHAADPRI 128
A+ DPRI
Sbjct 411 AYNNDPRI 418
> mmu:22791 Dnajc2, AU020218, MIDA1, Zrf1, Zrf2; DnaJ (Hsp40)
homolog, subfamily C, member 2; K09522 DnaJ homolog subfamily
C member 2
Length=621
Score = 112 bits (280), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 53/129 (41%), Positives = 85/129 (65%), Gaps = 3/129 (2%)
Query 1 RRQYDSALP-FDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
RR ++S P FD SVPS ++ + ++F+++F F+RN+RWS + V LGD ++ V
Sbjct 154 RRAFNSVDPTFDNSVPSKSEAK--DNFFQVFSPVFERNSRWSNKKNVPKLGDMNSSFEDV 211
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVE 119
+ FY FW+ F+SWR+F DE + ++A CR ER+W+E++N + + K E RI+ LV+
Sbjct 212 DAFYSFWYNFDSWREFSYLDEEEKEKAECRDERKWIEKQNRATRAQRKKEEMNRIRTLVD 271
Query 120 TAHAADPRI 128
A++ DPRI
Sbjct 272 NAYSCDPRI 280
> hsa:27000 DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1; DnaJ (Hsp40)
homolog, subfamily C, member 2; K09522 DnaJ homolog subfamily
C member 2
Length=568
Score = 112 bits (280), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/129 (41%), Positives = 85/129 (65%), Gaps = 3/129 (2%)
Query 1 RRQYDSALP-FDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
RR ++S P FD SVPS ++ + ++F+++F F+RN+RWS + V LGD ++ V
Sbjct 154 RRAFNSVDPTFDNSVPSKSEAK--DNFFEVFTPVFERNSRWSNKKNVPKLGDMNSSFEDV 211
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVE 119
+ FY FW+ F+SWR+F DE + ++A CR ERRW+E++N + + K E RI+ LV+
Sbjct 212 DIFYSFWYNFDSWREFSYLDEEEKEKAECRDERRWIEKQNRATRAQRKKEEMNRIRTLVD 271
Query 120 TAHAADPRI 128
A++ DPRI
Sbjct 272 NAYSCDPRI 280
> cel:F38A5.13 dnj-11; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-11); K09522 DnaJ homolog subfamily
C member 2
Length=589
Score = 92.0 bits (227), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 61/159 (38%), Positives = 92/159 (57%), Gaps = 2/159 (1%)
Query 1 RRQYDSA-LPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAV 59
R+ +DS F++ +P+ +FY FQ N+RWS +PV LG V
Sbjct 163 RQAFDSVDHKFNDIIPNEKSINH-NNFYNELAPVFQLNSRWSNIKPVPELGKSDATREDV 221
Query 60 EEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVE 119
E FY+FWF F+SWR+F DE D ++ R ERR ME++N +++ K E RI+KLV+
Sbjct 222 ENFYDFWFNFQSWREFSYLDEEDKERGEDRYERREMEKQNKAERERRRKEEAKRIRKLVD 281
Query 120 TAHAADPRIKQRKEEERKKKEEEKAAQLRAREEARRAAE 158
A+A DPRI + K+E++ KK++ K + RA E + A +
Sbjct 282 IAYAKDPRIIKFKKEQQAKKDKAKEDKQRAIREKQEAID 320
> sce:YGR285C ZUO1; Zuo1p; K09522 DnaJ homolog subfamily C member
2
Length=433
Score = 82.4 bits (202), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 56/132 (42%), Positives = 75/132 (56%), Gaps = 6/132 (4%)
Query 1 RRQYDSALPFDESVPSAADCQSPEDFYKIFGAAFQRNARWSVNRPVLTLGDCSTPLSAVE 60
R QYDS F VP + DFY+ +G F+ AR+S P+ +LG+ + VE
Sbjct 161 RAQYDSC-DFVADVPPPKKG-TDYDFYEAWGPVFEAEARFSKKTPIPSLGNKDSSKKEVE 218
Query 61 EFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMEREN--ARIKKKYIKNERARIQKLV 118
+FY FW F+SWR F DE D ++ R +R++ER+N AR KKK N AR+ KLV
Sbjct 219 QFYAFWHRFDSWRTFEFLDEDVPDDSSNRDHKRYIERKNKAARDKKKTADN--ARLVKLV 276
Query 119 ETAHAADPRIKQ 130
E A + DPRIK
Sbjct 277 ERAVSEDPRIKM 288
> tgo:TGME49_101340 DnaJ / zinc finger (C2H2 type) domain-containing
protein ; K09506 DnaJ homolog subfamily A member 5
Length=692
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 46/86 (53%), Gaps = 0/86 (0%)
Query 49 LGDCSTPLSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIK 108
LG ++P + V FY FW +F S + F D + R ERRW+++EN ++++ K
Sbjct 164 LGSSTSPWTEVAAFYSFWSSFASLKSFAFADSWKISPQDSRAERRWLQKENEKLRRAKRK 223
Query 109 NERARIQKLVETAHAADPRIKQRKEE 134
+Q+LV DPR+ QR +E
Sbjct 224 QFNDLVQRLVAAVKRRDPRVLQRSKE 249
> cpv:cgd5_2950 DNAj domain protein ; K09506 DnaJ homolog subfamily
A member 5
Length=588
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 51/101 (50%), Gaps = 3/101 (2%)
Query 49 LGDCSTPLSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIK 108
GD T L V EFY FW F S R F D +L+QA R RR ME EN + +KK K
Sbjct 180 FGDSKTNLDTVMEFYRFWSNFSSTRTFSWKDLWNLNQAQNRQIRRAMETENIKERKKGRK 239
Query 109 NERARIQKLVETAHAADPRI---KQRKEEERKKKEEEKAAQ 146
I+KL E DPR+ + K E+ K ++EK Q
Sbjct 240 EYNETIRKLTEYVKKRDPRVVSYMRNKSEQMIKLQKEKEMQ 280
> hsa:134218 DNAJC21, DNAJA5, GS3, JJJ1; DnaJ (Hsp40) homolog,
subfamily C, member 21; K09506 DnaJ homolog subfamily A member
5
Length=531
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 59/109 (54%), Gaps = 5/109 (4%)
Query 48 TLGDCSTPL-SAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKY 106
T GD + + V FY +W +F + ++F +E+D QA+ R E+R ME+EN +I+ K
Sbjct 136 TFGDSQSDYDTVVHPFYAYWQSFCTQKNFAWKEEYDTRQASNRWEKRAMEKENKKIRDKA 195
Query 107 IKNERARIQKLVETAHAADPRIKQRKEEERKKKEEEKAAQLRAREEARR 155
K + +++LV D R++ RK EE+ A + R EE RR
Sbjct 196 RKEKNELVRQLVAFIRKRDKRVQAH----RKLVEEQNAEKARKAEEMRR 240
> cel:T03F6.2 dnj-17; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-17); K09506 DnaJ homolog subfamily A
member 5
Length=510
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 55/112 (49%), Gaps = 8/112 (7%)
Query 49 LGDCSTPL-SAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYI 107
GD T L V FY FW +F + R F D +D+ QA+ R E R +++EN + +
Sbjct 160 FGDKDTDLEQTVNGFYGFWSSFSTTRSFAWLDHYDITQASNRFESRQIDQENKKFRDVGK 219
Query 108 KNERARIQKLVETAHAADPRIK-------QRKEEERKKKEEEKAAQLRAREE 152
+ +I+ LV DPR+K Q+K E KK+ + + Q+ +E
Sbjct 220 QERNEQIRNLVAFVRKRDPRVKAYREILEQKKLEAHKKQADNRKKQIAKNQE 271
> mmu:78244 Dnajc21, 4930461P20Rik, 9930116P15Rik; DnaJ (Hsp40)
homolog, subfamily C, member 21; K09506 DnaJ homolog subfamily
A member 5
Length=531
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 58/109 (53%), Gaps = 5/109 (4%)
Query 48 TLGDCSTPL-SAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKY 106
T GD + + V FY W +F + ++F +E+D QA+ R E+R ME+EN +I+ +
Sbjct 136 TFGDSQSDYDTVVHPFYAHWQSFCTQKNFSWKEEYDTRQASNRWEKRAMEKENKKIRDRA 195
Query 107 IKNERARIQKLVETAHAADPRIKQRKEEERKKKEEEKAAQLRAREEARR 155
K + +++LV D R++ RK EE+ A + R EE RR
Sbjct 196 RKEKNELVRQLVAFIRKRDKRVQAH----RKLVEEQNAEKARKAEEMRR 240
> ath:AT1G74250 DNAJ heat shock N-terminal domain-containing protein;
K09506 DnaJ homolog subfamily A member 5
Length=630
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 47/111 (42%), Gaps = 8/111 (7%)
Query 26 FYKIFGAAFQR--------NARWSVNRPVLTLGDCSTPLSAVEEFYEFWFAFESWRDFGV 77
+Y +F + + R R +G+ +P + V FY +W F + DF
Sbjct 127 YYDVFNSVYLNEIKFARTLGLRMDSVREAPIMGNLESPYAQVTAFYNYWLGFCTVMDFCW 186
Query 78 HDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQKLVETAHAADPRI 128
DE+D+ R RR ME EN + +KK + ++ L E D R+
Sbjct 187 VDEYDVMGGPNRKSRRMMEEENKKSRKKAKREYNDTVRGLAEFVKKRDKRV 237
> dre:336984 dnajc21, fa66c02, wu:fa66c02, zgc:63563; DnaJ (Hsp40)
homolog, subfamily C, member 21; K09506 DnaJ homolog subfamily
A member 5
Length=545
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/99 (32%), Positives = 53/99 (53%), Gaps = 4/99 (4%)
Query 57 SAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERARIQK 116
+ V FY +W +F + ++F +E+D QA+ R E+R ME+EN + + K K +++
Sbjct 148 TVVHLFYGYWQSFCTRKNFAWKEEYDTRQASNRWEKRAMEKENKKTRDKARKEHNELVRQ 207
Query 117 LVETAHAADPRIKQRKEEERKKKEEEKAAQLRAREEARR 155
LV D R++ K K EE+ A + + EE RR
Sbjct 208 LVAFVRKRDKRVQAHK----KLVEEQNAEKAKKVEELRR 242
> bbo:BBOV_IV006710 23.m06083; DnaJ domain containing protein;
K09506 DnaJ homolog subfamily A member 5
Length=341
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 58/107 (54%), Gaps = 11/107 (10%)
Query 49 LGDCSTPLSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMER----ENARIKK 104
G + V +FY+FW F + R F + +++ R+ RR++ER EN+++KK
Sbjct 155 FGTSQSTSQDVNKFYKFWHDFVTVRTFICEENWEIEG---RMNRRFVERQYKKENSKLKK 211
Query 105 KYIKNERARIQKLVETAHAADPRIKQRKEEERKKKEEEKAAQLRARE 151
+Y N ++ LV DPR+++ KEE+ + K ++ +L+ ++
Sbjct 212 EYNDN----VRNLVNIVKKVDPRVQRIKEEQNELKMLKELDKLKVQQ 254
> sce:YNL227C JJJ1; Co-chaperone that stimulates the ATPase activity
of Ssa1p, required for a late step of ribosome biogenesis;
associated with the cytosolic large ribosomal subunit;
contains a J-domain; mutation causes defects in fluid-phase
endocytosis; K09506 DnaJ homolog subfamily A member 5
Length=590
Score = 32.7 bits (73), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 49/105 (46%), Gaps = 3/105 (2%)
Query 49 LGDCSTPLSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIK 108
G T ++ FY+ W AF + + F DE+ + R +R + R N + +++
Sbjct 181 FGYSPTDYEYLKHFYKTWSAFNTLKSFSWKDEYMYSKNYDRRTKREVNRRNEKARQQARN 240
Query 109 NERARIQKLVETAHAADPRIKQR---KEEERKKKEEEKAAQLRAR 150
+++ V D R+K+ EE+RK KE+++ +L R
Sbjct 241 EYNKTVKRFVVFIKKLDKRMKEGAKIAEEQRKLKEQQRKNELNNR 285
> pfa:PF11_0486 MAEBL, putative
Length=2055
Score = 31.6 bits (70), Expect = 1.4, Method: Composition-based stats.
Identities = 33/121 (27%), Positives = 52/121 (42%), Gaps = 9/121 (7%)
Query 53 STPLSAVEEFYEFWFAFESWRDFGVHDEHDLDQAACRLERRWMERENARIKKKYIKNERA 112
S + +EE E+ + ++ +++E + ++ R+ R K Y +NE
Sbjct 1007 SVNIRKIEELTEYGNNDDVLKEKEINNEPIDNVNETKINRKSHVNSMERNKPSYKENEYD 1066
Query 113 RIQKLVETA---------HAADPRIKQRKEEERKKKEEEKAAQLRAREEARRAAEDQKRR 163
+++K VE A R EEE KKKE K A+ R E R AED +R
Sbjct 1067 QMEKNVEDETYSEEFGLFEEARKTETGRIEEESKKKEAMKRAEDARRIEEARRAEDARRI 1126
Query 164 E 164
E
Sbjct 1127 E 1127
Lambda K H
0.318 0.130 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3897828240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40