bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1204_orf2
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
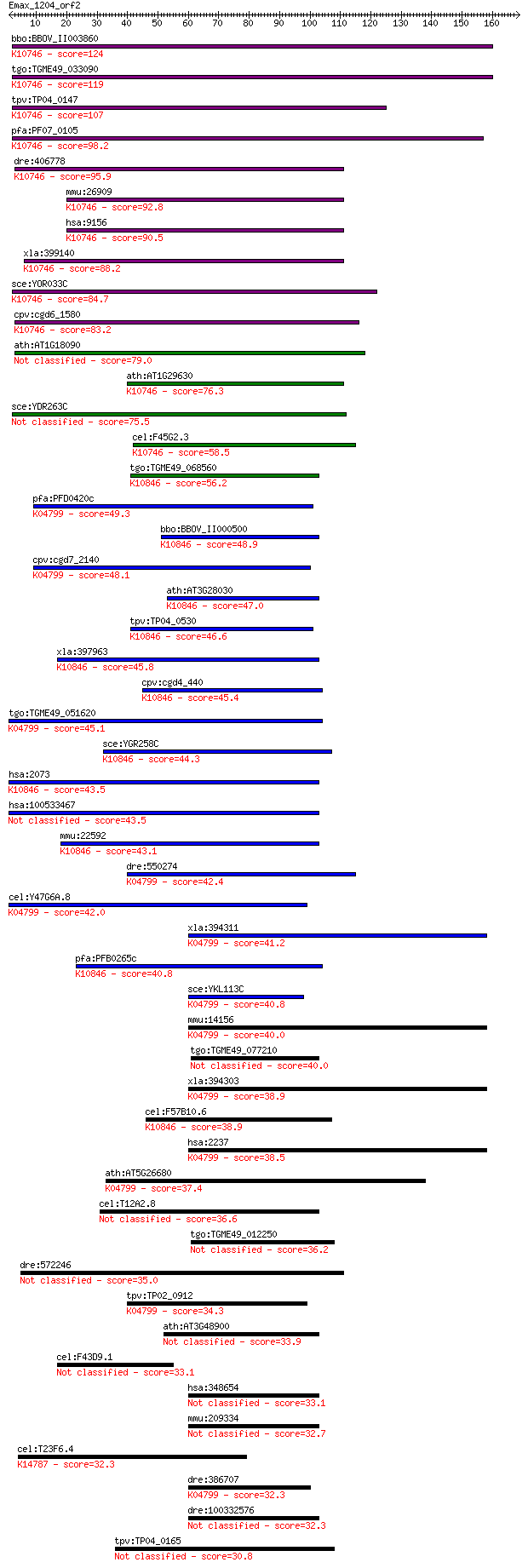
bbo:BBOV_II003860 18.m06322; XPG I-region family protein; K107... 124 2e-28
tgo:TGME49_033090 exonuclease, putative (EC:3.4.21.72); K10746... 119 5e-27
tpv:TP04_0147 exonuclease I; K10746 exonuclease 1 [EC:3.1.-.-] 107 1e-23
pfa:PF07_0105 exonuclease I, putative; K10746 exonuclease 1 [E... 98.2 1e-20
dre:406778 exo1, fc16e04, wu:fc16e04; zgc:55521; K10746 exonuc... 95.9 5e-20
mmu:26909 Exo1, 5730442G03Rik, Msa; exonuclease 1; K10746 exon... 92.8 5e-19
hsa:9156 EXO1, HEX1, hExoI; exonuclease 1; K10746 exonuclease ... 90.5 3e-18
xla:399140 exo1, MGC83785, exoi; exonuclease 1; K10746 exonucl... 88.2 1e-17
sce:YOR033C EXO1, DHS1; 5'-3' exonuclease and flap-endonucleas... 84.7 1e-16
cpv:cgd6_1580 exonuclease i/din7p-like; xeroderma pigmentosum ... 83.2 3e-16
ath:AT1G18090 exonuclease, putative 79.0 7e-15
ath:AT1G29630 DNA binding / catalytic/ nuclease; K10746 exonuc... 76.3 4e-14
sce:YDR263C DIN7, DIN3; Din7p 75.5 7e-14
cel:F45G2.3 hypothetical protein; K10746 exonuclease 1 [EC:3.1... 58.5 9e-09
tgo:TGME49_068560 RAD2 endonuclease, putative ; K10846 DNA exc... 56.2 5e-08
pfa:PFD0420c FEN-1, PfFEN-1; flap endonuclease 1; K04799 flap ... 49.3 6e-06
bbo:BBOV_II000500 18.m06025; Rad2 endonuclease; K10846 DNA exc... 48.9 8e-06
cpv:cgd7_2140 flap endonuclease 1 ; K04799 flap endonuclease-1... 48.1 1e-05
ath:AT3G28030 UVH3; UVH3 (ULTRAVIOLET HYPERSENSITIVE 3); DNA b... 47.0 3e-05
tpv:TP04_0530 DNA repair protein Rad2; K10846 DNA excision rep... 46.6 4e-05
xla:397963 ercc5, cofs3, ercm2, uvdr, xpg, xpgc; excision repa... 45.8 7e-05
cpv:cgd4_440 XPG, DNA excision repair protein, flap endonuclea... 45.4 1e-04
tgo:TGME49_051620 flap endonuclease-1, putative ; K04799 flap ... 45.1 1e-04
sce:YGR258C RAD2; Rad2p; K10846 DNA excision repair protein ER... 44.3 2e-04
hsa:2073 ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC; excision repair... 43.5 3e-04
hsa:100533467 BIVM-ERCC5; BIVM-ERCC5 readthrough 43.5 4e-04
mmu:22592 Ercc5, MGC176031, Xpg; excision repair cross-complem... 43.1 4e-04
dre:550274 im:7147072; zgc:110269; K04799 flap endonuclease-1 ... 42.4 7e-04
cel:Y47G6A.8 crn-1; Cell-death-Related Nuclease family member ... 42.0 0.001
xla:394311 fen1-b, fen-1, mf1, rad2; flap structure-specific e... 41.2 0.002
pfa:PFB0265c DNA repair endonuclease, putative; K10846 DNA exc... 40.8 0.002
sce:YKL113C RAD27, ERC11, FEN1, RTH1; 5' to 3' exonuclease, 5'... 40.8 0.002
mmu:14156 Fen1, AW538437; flap structure specific endonuclease... 40.0 0.003
tgo:TGME49_077210 XPG N-terminal domain containing protein 40.0 0.004
xla:394303 fen1-a, MGC196488, MGC196492, fen-1, fen1, mf1, rad... 38.9 0.008
cel:F57B10.6 xpg-1; XPG (Xeroderma Pigmentosum group G) DNA re... 38.9 0.009
hsa:2237 FEN1, FEN-1, MF1, RAD2; flap structure-specific endon... 38.5 0.010
ath:AT5G26680 endonuclease, putative; K04799 flap endonuclease... 37.4 0.022
cel:T12A2.8 hypothetical protein 36.6 0.044
tgo:TGME49_012250 XPG N-terminal domain containing protein 36.2 0.060
dre:572246 TPR repeat-containing protein YDR161W-like 35.0 0.12
tpv:TP02_0912 flap endonuclease 1; K04799 flap endonuclease-1 ... 34.3 0.20
ath:AT3G48900 DNA binding / catalytic/ chromatin binding / nuc... 33.9 0.28
cel:F43D9.1 hypothetical protein 33.1 0.40
hsa:348654 GEN1, DKFZp781F0986, FLJ40869, Gen; Gen homolog 1, ... 33.1 0.42
mmu:209334 Gen1, 5830483C08Rik, MGC115970; Gen homolog 1, endo... 32.7 0.65
cel:T23F6.4 rbd-1; RBD (RNA binding domain) protein family mem... 32.3 0.67
dre:386707 fen1, cb879; flap structure-specific endonuclease 1... 32.3 0.82
dre:100332576 Gen homolog 1, endonuclease-like 32.3 0.87
tpv:TP04_0165 hypothetical protein 30.8 2.0
> bbo:BBOV_II003860 18.m06322; XPG I-region family protein; K10746
exonuclease 1 [EC:3.1.-.-]
Length=501
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 63/158 (39%), Positives = 100/158 (63%), Gaps = 19/158 (12%)
Query 2 EDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELG 61
E++ R++RR+ AR+EAL ++++ + E+M KC Q+I ITP ++ +V+ CR +
Sbjct 100 ENQLRRERRDKAREEALAMIEKNGGAINT---EIMRKCMQAIHITPEVIARVMEICRAMN 156
Query 62 IRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPFLQ 121
+R +VAP+EADAQ++YL R+ ++A+SEDSDLL YGC RV K++++GK + L L F +
Sbjct 157 VRIVVAPYEADAQVSYLCRSGIAYAALSEDSDLLVYGCPRVWFKLERDGKADELTLGFNK 216
Query 122 DENDNCISELQAHLNKTAKQMELLRILKGLNHERFVAM 159
D + C + L LKGL+H F+AM
Sbjct 217 DPDVKCNTGL----------------LKGLSHRMFIAM 238
> tgo:TGME49_033090 exonuclease, putative (EC:3.4.21.72); K10746
exonuclease 1 [EC:3.1.-.-]
Length=951
Score = 119 bits (298), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 71/160 (44%), Positives = 98/160 (61%), Gaps = 16/160 (10%)
Query 2 EDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELG 61
EDEKRQ+ R+ A +EA ELLK+ Y+E + T VD VI+ACR LG
Sbjct 110 EDEKRQQARQKASEEARELLKK--------YEEARRAGRKPPGDT--RVDTVISACRSLG 159
Query 62 IRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPFLQ 121
+ +VAP+EADAQLA+L+RT +I +A+SEDSDLLA+GC++V+ KMD+EG CE L LP
Sbjct 160 VAFVVAPYEADAQLAFLARTGKIAAAVSEDSDLLAHGCQQVLFKMDREGNCERLSLPL-- 217
Query 122 DENDNCISE--LQAHLNKTAKQMELLRILKGLNHERFVAM 159
ND + + +AK++ L L+ + F AM
Sbjct 218 --NDRASPDAAQASVSASSAKKLGQLECLRDFDQTMFTAM 255
> tpv:TP04_0147 exonuclease I; K10746 exonuclease 1 [EC:3.1.-.-]
Length=550
Score = 107 bits (268), Expect = 1e-23, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 84/123 (68%), Gaps = 3/123 (2%)
Query 2 EDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELG 61
E+ R++RR AR EALE++++ + + E+M KC Q+I ITP +V +VI C+++
Sbjct 99 ENSIRRERRNKARAEALEMIRKNKGKINT---EIMRKCMQAIQITPEIVHRVITICKKVN 155
Query 62 IRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPFLQ 121
+ +V+P+EADAQ++YL RT AISEDSDL+ YGC +++ K++KEGK L +PF
Sbjct 156 VSVVVSPYEADAQISYLCRTGIADFAISEDSDLIVYGCPKIIFKLNKEGKGVELNVPFFN 215
Query 122 DEN 124
+N
Sbjct 216 KQN 218
> pfa:PF07_0105 exonuclease I, putative; K10746 exonuclease 1
[EC:3.1.-.-]
Length=1347
Score = 98.2 bits (243), Expect = 1e-20, Method: Composition-based stats.
Identities = 57/156 (36%), Positives = 92/156 (58%), Gaps = 3/156 (1%)
Query 2 EDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELG 61
E+ R+ RRE A+ E E++ + + P + ++ KC Q+IS++ ++D V CR+
Sbjct 88 ENMIRKNRREKAKMELQEIISKVKN--PRTNEMVLKKCIQAISVSKEIIDSVKEFCRKKN 145
Query 62 IRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPFLQ 121
I I++P+EADAQL+YL R I AISEDSDLL YGC RV+ K+ G+C + L +
Sbjct 146 IDYIISPYEADAQLSYLCRMGFISCAISEDSDLLVYGCPRVLYKLKNTGECNEISLMPIN 205
Query 122 DEND-NCISELQAHLNKTAKQMELLRILKGLNHERF 156
D D N I++++ L+ + + + I K N + +
Sbjct 206 DLIDWNVINKIKNPLSNSYNEFYITPIKKLQNSDDY 241
> dre:406778 exo1, fc16e04, wu:fc16e04; zgc:55521; K10746 exonuclease
1 [EC:3.1.-.-]
Length=806
Score = 95.9 bits (237), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 47/108 (43%), Positives = 75/108 (69%), Gaps = 6/108 (5%)
Query 3 DEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGI 62
++ R++RR+ Q+ +LL+ + E ++S++ITP+M VI A R G+
Sbjct 89 EKSRRERRQANLQKGKQLLREGK------ITEARECFTRSVNITPSMAHDVIRAARTRGV 142
Query 63 RCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEG 110
C+VAP+EADAQLA+L++++ + I+EDSDLLA+GCK+V+LKMDK+G
Sbjct 143 DCVVAPYEADAQLAFLNKSDIAQAVITEDSDLLAFGCKKVILKMDKQG 190
> mmu:26909 Exo1, 5730442G03Rik, Msa; exonuclease 1; K10746 exonuclease
1 [EC:3.1.-.-]
Length=837
Score = 92.8 bits (229), Expect = 5e-19, Method: Composition-based stats.
Identities = 49/91 (53%), Positives = 64/91 (70%), Gaps = 0/91 (0%)
Query 20 LLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLS 79
LLK KQ E ++SI+IT AM KVI A R LG+ C+VAP+EADAQLAYL+
Sbjct 100 LLKGKQLLREGKVSEARDCFARSINITHAMAHKVIKAARALGVDCLVAPYEADAQLAYLN 159
Query 80 RTNQIHSAISEDSDLLAYGCKRVMLKMDKEG 110
+ + + I+EDSDLLA+GCK+V+LKMD+ G
Sbjct 160 KAGIVQAVITEDSDLLAFGCKKVILKMDQFG 190
> hsa:9156 EXO1, HEX1, hExoI; exonuclease 1; K10746 exonuclease
1 [EC:3.1.-.-]
Length=803
Score = 90.5 bits (223), Expect = 3e-18, Method: Composition-based stats.
Identities = 48/91 (52%), Positives = 63/91 (69%), Gaps = 0/91 (0%)
Query 20 LLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLS 79
LLK KQ E ++SI+IT AM KVI A R G+ C+VAP+EADAQLAYL+
Sbjct 100 LLKGKQLLREGKVSEARECFTRSINITHAMAHKVIKAARSQGVDCLVAPYEADAQLAYLN 159
Query 80 RTNQIHSAISEDSDLLAYGCKRVMLKMDKEG 110
+ + + I+EDSDLLA+GCK+V+LKMD+ G
Sbjct 160 KAGIVQAIITEDSDLLAFGCKKVILKMDQFG 190
> xla:399140 exo1, MGC83785, exoi; exonuclease 1; K10746 exonuclease
1 [EC:3.1.-.-]
Length=734
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/105 (44%), Positives = 71/105 (67%), Gaps = 6/105 (5%)
Query 6 RQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGIRCI 65
R+++R+T Q+ +LL+ + E S+S++IT +M +VI A R G+ I
Sbjct 92 RREKRQTNLQKGKQLLREGK------LAEARECFSRSVNITSSMAHEVIKAARSEGVDYI 145
Query 66 VAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEG 110
VAP+EAD+QLAYL++ + + I+EDSDLLA+GCK+V+LKMDK G
Sbjct 146 VAPYEADSQLAYLNKNDFAEAIITEDSDLLAFGCKKVLLKMDKFG 190
> sce:YOR033C EXO1, DHS1; 5'-3' exonuclease and flap-endonuclease
involved in recombination, double-strand break repair and
DNA mismatch repair; member of the Rad2p nuclease family,
with conserved N and I nuclease domains (EC:3.1.-.-); K10746
exonuclease 1 [EC:3.1.-.-]
Length=702
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 51/122 (41%), Positives = 73/122 (59%), Gaps = 10/122 (8%)
Query 2 EDEKRQKRRET-ARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACREL 60
E ++R KR+E A E L K+ + DY KC + ITP M +I C+
Sbjct 89 ESKRRDKRKENKAIAERLWACGEKKNAM--DY---FQKC---VDITPEMAKCIICYCKLN 140
Query 61 GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKC-EVLRLPF 119
GIR IVAPFEAD+Q+ YL + N + ISEDSDLL +GC+R++ K++ G+C E+ R F
Sbjct 141 GIRYIVAPFEADSQMVYLEQKNIVQGIISEDSDLLVFGCRRLITKLNDYGECLEICRDNF 200
Query 120 LQ 121
++
Sbjct 201 IK 202
> cpv:cgd6_1580 exonuclease i/din7p-like; xeroderma pigmentosum
G N-region plus xeroderma pigmentosum G I-region plus HhH2
domain ; K10746 exonuclease 1 [EC:3.1.-.-]
Length=482
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 47/115 (40%), Positives = 69/115 (60%), Gaps = 5/115 (4%)
Query 3 DEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACR-ELG 61
+E+R KRR A++E + L K+ + + L C +++ ITP + +V+ R E
Sbjct 89 EEERSKRRSDAKREIIRLKSEKKSYSSYNMRSL---CQKALDITPNIAHQVLEVLRDEYK 145
Query 62 IRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKM-DKEGKCEVL 115
I CIVAP+EADAQL+YLSR I + I+EDSD+L +G + K DK G C V+
Sbjct 146 IECIVAPYEADAQLSYLSRIKYIDAVITEDSDMLVFGSICTIYKYDDKTGNCRVI 200
> ath:AT1G18090 exonuclease, putative
Length=577
Score = 79.0 bits (193), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 51/119 (42%), Positives = 73/119 (61%), Gaps = 11/119 (9%)
Query 3 DEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGI 62
DE+ +KR+ A +A ++K K+ N+ EL + ++S+T +M ++I + +
Sbjct 93 DERHRKRK--ANFDA-AMVKLKEGNVAA-ATELFQR---AVSVTSSMAHQLIQVLKSENV 145
Query 63 RCIVAPFEADAQLAYLSRT----NQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRL 117
IVAP+EADAQLAYLS I + I+EDSDLLAYGCK V+ KMD+ GK E L L
Sbjct 146 EFIVAPYEADAQLAYLSSLELEQGGIAAVITEDSDLLAYGCKAVIFKMDRYGKGEELVL 204
> ath:AT1G29630 DNA binding / catalytic/ nuclease; K10746 exonuclease
1 [EC:3.1.-.-]
Length=735
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 53/71 (74%), Gaps = 0/71 (0%)
Query 40 SQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGC 99
S+++ I+P++ ++I R+ + +VAP+EADAQ+A+L+ T Q+ + I+EDSDL+ +GC
Sbjct 120 SKAVDISPSIAHELIQVLRQENVDYVVAPYEADAQMAFLAITKQVDAIITEDSDLIPFGC 179
Query 100 KRVMLKMDKEG 110
R++ KMDK G
Sbjct 180 LRIIFKMDKFG 190
> sce:YDR263C DIN7, DIN3; Din7p
Length=430
Score = 75.5 bits (184), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 61/110 (55%), Gaps = 7/110 (6%)
Query 2 EDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELG 61
E +R+KR E E++ +K + Y M +S+ ITP M +I C+
Sbjct 89 ETRRRKKRLEN------EMIAKKLWSAGNRYNA-MEYFQKSVDITPEMAKCIIDYCKLHS 141
Query 62 IRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGK 111
I IVAPFEAD Q+ YL + I ISEDSDLL +GCK ++ K++ +GK
Sbjct 142 IPYIVAPFEADPQMVYLEKMGLIQGIISEDSDLLVFGCKTLITKLNDQGK 191
> cel:F45G2.3 hypothetical protein; K10746 exonuclease 1 [EC:3.1.-.-]
Length=639
Score = 58.5 bits (140), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 46/75 (61%), Gaps = 2/75 (2%)
Query 42 SISITPAMVDKVIAACREL-GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCK 100
+ SI+ +V+ I R + + +VAP+EADAQLAYL + + + I+EDSDL+ +GC+
Sbjct 122 ATSISADIVENTIQYFRSMTNVDIVVAPYEADAQLAYLVQEKLVDAVITEDSDLIVFGCE 181
Query 101 RVMLKM-DKEGKCEV 114
+ K G+C V
Sbjct 182 MIYFKWQSATGECSV 196
> tgo:TGME49_068560 RAD2 endonuclease, putative ; K10846 DNA excision
repair protein ERCC-5
Length=2004
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 39/62 (62%), Gaps = 0/62 (0%)
Query 41 QSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCK 100
Q+ +IT M D+VIA R G+ I AP EA+A AYL++ N + IS+DSD L +G +
Sbjct 1417 QTGNITERMKDQVIALLRAFGVPFITAPGEAEATAAYLTKQNLADAVISDDSDALVFGAR 1476
Query 101 RV 102
+
Sbjct 1477 EI 1478
> pfa:PFD0420c FEN-1, PfFEN-1; flap endonuclease 1; K04799 flap
endonuclease-1 [EC:3.-.-.-]
Length=672
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 53/92 (57%), Gaps = 2/92 (2%)
Query 9 RRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGIRCIVAP 68
+R RQ+A ELLK+ ++ + +E+ + +++ +T ++ +GI I AP
Sbjct 101 KRGEKRQKAEELLKKAKE--EGNLEEIKKQSGRTVRVTRKQNEEAKKLLTLMGIPIIEAP 158
Query 69 FEADAQLAYLSRTNQIHSAISEDSDLLAYGCK 100
EA++Q A+L++ N H+ +ED+D L +G K
Sbjct 159 CEAESQCAFLTKYNLAHATATEDADALVFGTK 190
> bbo:BBOV_II000500 18.m06025; Rad2 endonuclease; K10846 DNA excision
repair protein ERCC-5
Length=1002
Score = 48.9 bits (115), Expect = 8e-06, Method: Composition-based stats.
Identities = 27/52 (51%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 51 DKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
DKV A G+ IVAP EA+AQ A+L+ T IS+DSD LA+G KRV
Sbjct 687 DKVPALLDLFGVPFIVAPSEAEAQCAHLNNTGLCFGVISDDSDTLAFGAKRV 738
> cpv:cgd7_2140 flap endonuclease 1 ; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=490
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 54/92 (58%), Gaps = 4/92 (4%)
Query 9 RRETARQEALELLKRKQQNLPVDYKELMSKCS-QSISITPAMVDKVIAACRELGIRCIVA 67
+R+ R++AL L++ Q+ + +EL+ K S ++I +T V+ V LG+ CI A
Sbjct 101 KRDERREKALAELEKAQE---IGDEELIKKQSVRTIHVTKKQVEDVKKLLGFLGMPCIDA 157
Query 68 PFEADAQLAYLSRTNQIHSAISEDSDLLAYGC 99
P EA+AQ A L + ++ ++ED+D L +G
Sbjct 158 PSEAEAQCAELCKDGLVYGVVTEDADSLTFGT 189
> ath:AT3G28030 UVH3; UVH3 (ULTRAVIOLET HYPERSENSITIVE 3); DNA
binding / catalytic/ endonuclease/ nuclease/ single-stranded
DNA binding; K10846 DNA excision repair protein ERCC-5
Length=1479
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 35/54 (64%), Gaps = 4/54 (7%)
Query 53 VIAACREL----GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
+ A C+EL GI I+AP EA+AQ A++ ++N + +++DSD+ +G + V
Sbjct 925 MFAECQELLQIFGIPYIIAPMEAEAQCAFMEQSNLVDGIVTDDSDVFLFGARSV 978
> tpv:TP04_0530 DNA repair protein Rad2; K10846 DNA excision repair
protein ERCC-5
Length=835
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/62 (40%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 41 QSISIT--PAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYG 98
QS+S+T D V G+ IVAP EA++Q AY++R+ + ++ IS+DSD L +G
Sbjct 517 QSVSVTHKNPYYDDVHKLLEHFGVPYIVAPSEAESQCAYMNRSGKCYAVISDDSDSLVFG 576
Query 99 CK 100
K
Sbjct 577 AK 578
> xla:397963 ercc5, cofs3, ercm2, uvdr, xpg, xpgc; excision repair
cross-complementing rodent repair deficiency, complementation
group 5; K10846 DNA excision repair protein ERCC-5
Length=1197
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 47/90 (52%), Gaps = 4/90 (4%)
Query 17 ALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACREL----GIRCIVAPFEAD 72
++E L+ + NL V L ++ Q I + ++ +EL GI IVAP EA+
Sbjct 766 SVEELESLENNLYVQQTSLQAQRQQQERIAATVTGQMCLESQELLQLFGIPYIVAPMEAE 825
Query 73 AQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
AQ A L T+Q I++DSD+ +G + V
Sbjct 826 AQCAILDLTDQTSGTITDDSDIWLFGARHV 855
> cpv:cgd4_440 XPG, DNA excision repair protein, flap endonuclease
; K10846 DNA excision repair protein ERCC-5
Length=1147
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 35/59 (59%), Gaps = 0/59 (0%)
Query 45 ITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVM 103
IT M ++ + LGI I +P EA+AQ + L++ N H +S+DSD L +G K++
Sbjct 875 ITKEMQYQICLLLKALGIPWIDSPGEAEAQASILTQLNICHGVLSDDSDCLIFGAKKIF 933
> tgo:TGME49_051620 flap endonuclease-1, putative ; K04799 flap
endonuclease-1 [EC:3.-.-.-]
Length=552
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/103 (32%), Positives = 60/103 (58%), Gaps = 6/103 (5%)
Query 1 GEDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACREL 60
GE KR++ RE+A QEA E R++ N+ +EL + +S+ ++ + V R +
Sbjct 97 GELAKRRELRESA-QEAAEKA-REEGNV----EELRKQIVRSVRVSKQHNEDVKRLLRLM 150
Query 61 GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVM 103
G+ + AP EA+AQ A L++ ++ + +ED+D L +G R++
Sbjct 151 GLPVVEAPCEAEAQCAELTKNRKVWATATEDADALTFGATRLI 193
> sce:YGR258C RAD2; Rad2p; K10846 DNA excision repair protein
ERCC-5
Length=1031
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 32 YKELMSKCSQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISED 91
+++ M S +T M+ +V GI I AP EA+AQ A L + N + I++D
Sbjct 754 FEQQMKDKRDSDEVTMDMIKEVQELLSRFGIPYITAPMEAEAQCAELLQLNLVDGIITDD 813
Query 92 SDLLAYGCKRVMLKM 106
SD+ +G ++ M
Sbjct 814 SDVFLFGGTKIYKNM 828
> hsa:2073 ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC; excision repair
cross-complementing rodent repair deficiency, complementation
group 5; K10846 DNA excision repair protein ERCC-5
Length=1186
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 53/110 (48%), Gaps = 8/110 (7%)
Query 1 GEDEKRQKRRETARQE----ALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAA 56
GE ++ +K E + E LE L+ + NL L ++ Q I + ++
Sbjct 712 GEPQEAEKDAEDSLHEWQDINLEELETLESNLLAQQNSLKAQKQQQERIAATVTGQMFLE 771
Query 57 CREL----GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
+EL GI I AP EA+AQ A L T+Q I++DSD+ +G + V
Sbjct 772 SQELLRLFGIPYIQAPMEAEAQCAILDLTDQTSGTITDDSDIWLFGARHV 821
> hsa:100533467 BIVM-ERCC5; BIVM-ERCC5 readthrough
Length=1640
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 53/110 (48%), Gaps = 8/110 (7%)
Query 1 GEDEKRQKRRETARQE----ALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAA 56
GE ++ +K E + E LE L+ + NL L ++ Q I + ++
Sbjct 1166 GEPQEAEKDAEDSLHEWQDINLEELETLESNLLAQQNSLKAQKQQQERIAATVTGQMFLE 1225
Query 57 CREL----GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
+EL GI I AP EA+AQ A L T+Q I++DSD+ +G + V
Sbjct 1226 SQELLRLFGIPYIQAPMEAEAQCAILDLTDQTSGTITDDSDIWLFGARHV 1275
> mmu:22592 Ercc5, MGC176031, Xpg; excision repair cross-complementing
rodent repair deficiency, complementation group 5;
K10846 DNA excision repair protein ERCC-5
Length=1170
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 45/89 (50%), Gaps = 4/89 (4%)
Query 18 LELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACREL----GIRCIVAPFEADA 73
LE L + NL + L ++ Q I ++ ++ +EL G+ I AP EA+A
Sbjct 732 LEELDALESNLLAEQNSLKAQKQQQDRIAASVTGQMFLESQELLRLFGVPYIQAPMEAEA 791
Query 74 QLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
Q A L T+Q I++DSD+ +G + V
Sbjct 792 QCAMLDLTDQTSGTITDDSDIWLFGARHV 820
> dre:550274 im:7147072; zgc:110269; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=350
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 42/75 (56%), Gaps = 0/75 (0%)
Query 40 SQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGC 99
SQS + A + + +G+ CI AP EA+A A+L++ +++ SED D LA+G
Sbjct 100 SQSPNTGSAFNQECLRLLHLMGVPCIKAPGEAEALCAHLAKIGTVNAVASEDMDTLAFGG 159
Query 100 KRVMLKMDKEGKCEV 114
++ +++ + E+
Sbjct 160 TVLLRQLNAKRDSEI 174
> cel:Y47G6A.8 crn-1; Cell-death-Related Nuclease family member
(crn-1); K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 49/98 (50%), Gaps = 6/98 (6%)
Query 1 GEDEKRQKRRETARQEALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACREL 60
GE EKR +RR A ++AL K K D KE + + +T D+ +
Sbjct 95 GELEKRSERRAEA-EKALTEAKEKG-----DVKEAEKFERRLVKVTKQQNDEAKRLLGLM 148
Query 61 GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYG 98
GI + AP EA+AQ A+L + ++ ++ED D L +G
Sbjct 149 GIPVVEAPCEAEAQCAHLVKAGKVFGTVTEDMDALTFG 186
> xla:394311 fen1-b, fen-1, mf1, rad2; flap structure-specific
endonuclease 1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 21/98 (21%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPF 119
+GI + AP EA+A A L + ++++A +ED D L +G ++ + E +LP
Sbjct 148 MGIPYVDAPCEAEATCAALVKAGKVYAAATEDMDALTFGTPVLLRHLT---ASEAKKLP- 203
Query 120 LQDENDNCISELQAHLNKTAKQMELLRILKGLNHERFV 157
I E HLN+ + + G+NHE+FV
Sbjct 204 --------IQEF--HLNRVFQDI-------GINHEQFV 224
> pfa:PFB0265c DNA repair endonuclease, putative; K10846 DNA excision
repair protein ERCC-5
Length=1516
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 3/81 (3%)
Query 23 RKQQNLPVDYKELMSKCSQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTN 82
++ + L +YK+L +I I M D + GI I +P EA+AQ +YL+ N
Sbjct 1188 KENEELIKEYKKLKK---NNIEINDEMNDDIKLLLNFFGIPYIQSPCEAEAQCSYLNNKN 1244
Query 83 QIHSAISEDSDLLAYGCKRVM 103
+ IS+DSD+L + K V+
Sbjct 1245 YCDAIISDDSDVLVFSGKTVI 1265
> sce:YKL113C RAD27, ERC11, FEN1, RTH1; 5' to 3' exonuclease,
5' flap endonuclease, required for Okazaki fragment processing
and maturation as well as for long-patch base-excision repair;
member of the S. pombe RAD2/FEN1 family; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=382
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 27/38 (71%), Gaps = 0/38 (0%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAY 97
+GI I+AP EA+AQ A L++ ++++A SED D L Y
Sbjct 146 MGIPYIIAPTEAEAQCAELAKKGKVYAAASEDMDTLCY 183
> mmu:14156 Fen1, AW538437; flap structure specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=380
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 49/98 (50%), Gaps = 21/98 (21%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPF 119
+GI + AP EA+A A L++ ++++A +ED D L +G +M + E +LP
Sbjct 148 MGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLT---ASEAKKLP- 203
Query 120 LQDENDNCISELQAHLNKTAKQMELLRILKGLNHERFV 157
I E HL++ +++ GLN E+FV
Sbjct 204 --------IQEF--HLSRVLQEL-------GLNQEQFV 224
> tgo:TGME49_077210 XPG N-terminal domain containing protein
Length=268
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 61 GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
G+ CI A +EADAQ+A L + ++ED DLLAY + V
Sbjct 171 GVVCISAAYEADAQMARLVADGFADAVLTEDGDLLAYQARMV 212
> xla:394303 fen1-a, MGC196488, MGC196492, fen-1, fen1, mf1, rad2,
xFEN1a; flap structure-specific endonuclease 1; K04799
flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 47/98 (47%), Gaps = 21/98 (21%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPF 119
+GI + AP EA+A A L + ++++A +ED D L +G ++ + E +LP
Sbjct 148 MGIPYVDAPCEAEATCAALVKAGKVYAAATEDMDALTFGTPVLLRHLT---ASEAKKLP- 203
Query 120 LQDENDNCISELQAHLNKTAKQMELLRILKGLNHERFV 157
I E HLN+ + + G+ HE+FV
Sbjct 204 --------IQEF--HLNRVIQDI-------GITHEQFV 224
> cel:F57B10.6 xpg-1; XPG (Xeroderma Pigmentosum group G) DNA
repair gene homolog family member (xpg-1); K10846 DNA excision
repair protein ERCC-5
Length=829
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 46 TPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLK 105
TP + + GI I +P EA+AQ L R + +S+DSD+ A+G + V
Sbjct 474 TPELYRDLQEFLTNAGIPWIESPGEAEAQCVELERLGLVDGVVSDDSDVWAFGAQHVYRH 533
Query 106 M 106
M
Sbjct 534 M 534
> hsa:2237 FEN1, FEN-1, MF1, RAD2; flap structure-specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=380
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 21/98 (21%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEGKCEVLRLPF 119
+GI + AP EA+A A L + ++++A +ED D L +G +M + E +LP
Sbjct 148 MGIPYLDAPSEAEASCAALVKAGKVYAAATEDMDCLTFGSPVLMRHLT---ASEAKKLP- 203
Query 120 LQDENDNCISELQAHLNKTAKQMELLRILKGLNHERFV 157
I E HL++ +++ GLN E+FV
Sbjct 204 --------IQEF--HLSRILQEL-------GLNQEQFV 224
> ath:AT5G26680 endonuclease, putative; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=383
Score = 37.4 bits (85), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 54/107 (50%), Gaps = 6/107 (5%)
Query 33 KELMSKCSQ-SISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISED 91
KE + K S+ ++ +T D R +G+ + A EA+AQ A L ++ +++ SED
Sbjct 121 KEDIEKYSKRTVKVTKQHNDDCKRLLRLMGVPVVEATSEAEAQCAALCKSGKVYGVASED 180
Query 92 SDLLAYGCKRVMLK-MDKEGKCEVLRLPFLQDENDNCISELQAHLNK 137
D L +G + + MD + ++P ++ E + ELQ +++
Sbjct 181 MDSLTFGAPKFLRHLMDPSSR----KIPVMEFEVAKILEELQLTMDQ 223
> cel:T12A2.8 hypothetical protein
Length=434
Score = 36.6 bits (83), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 36/77 (46%), Gaps = 5/77 (6%)
Query 31 DYKELMSKCSQSISITPA--MVDKVI---AACRELGIRCIVAPFEADAQLAYLSRTNQIH 85
D E + + +S +P +VD V A ELGI+ I+AP + +AQ A L
Sbjct 89 DQNEFVPRKRRSFGDSPFTNLVDHVYKTNALLTELGIKVIIAPGDGEAQCARLEDLGVTS 148
Query 86 SAISEDSDLLAYGCKRV 102
I+ D D +G K +
Sbjct 149 GCITTDFDYFLFGGKNL 165
> tgo:TGME49_012250 XPG N-terminal domain containing protein
Length=767
Score = 36.2 bits (82), Expect = 0.060, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 61 GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMD 107
G C APF A AQLAY + N + AI LL +G RV++ +D
Sbjct 141 GCECFQAPFFATAQLAYFAEQNFL-DAIFGPPSLLLFGVPRVIVNID 186
> dre:572246 TPR repeat-containing protein YDR161W-like
Length=401
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 26/110 (23%), Positives = 55/110 (50%), Gaps = 8/110 (7%)
Query 5 KRQKRRETARQEALELLKRKQQNLP-VDYKELMSKCSQSISITPA---MVDKVIAACREL 60
+R K++ + +LL++ ++++ D+ C +++ I P ++D + + C EL
Sbjct 38 ERAKKKTAEKYTVGQLLQKTEESVDNFDFDMARLYCQRALDIEPTNLTILDMMGSICSEL 97
Query 61 GIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRVMLKMDKEG 110
G ++P E ++ YL QIH+ + E + G + ++ MDK G
Sbjct 98 GDAVELSPEEGHSKYMYL---GQIHTGM-EAVQYFSKGIEIMLNTMDKHG 143
> tpv:TP02_0912 flap endonuclease 1; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=494
Score = 34.3 bits (77), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 0/59 (0%)
Query 40 SQSISITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYG 98
+++ ++ M + R +G+ I A EA+AQ AYL N SED+D L +G
Sbjct 130 GRTVKVSKEMNESAKKLLRLMGVPVIEALEEAEAQCAYLVTKNLCRFVASEDTDTLVFG 188
> ath:AT3G48900 DNA binding / catalytic/ chromatin binding / nuclease
Length=600
Score = 33.9 bits (76), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query 52 KVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
KVIA+ LGI C+ EA+AQ A L+ + + S DSD+ +G K V
Sbjct 126 KVIAS--TLGILCLDGIEEAEAQCALLNSESLCDACFSFDSDIFLFGAKTV 174
> cel:F43D9.1 hypothetical protein
Length=1226
Score = 33.1 bits (74), Expect = 0.40, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 17 ALELLKRKQQNLPVDYKELMSKCSQSISITPAMVDKVI 54
A LK+K + PV+ +E M++CS MVD++I
Sbjct 306 AYTALKKKLDHFPVEKREFMTQCSLECETDKDMVDRII 343
> hsa:348654 GEN1, DKFZp781F0986, FLJ40869, Gen; Gen homolog 1,
endonuclease (Drosophila)
Length=908
Score = 33.1 bits (74), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
LGI + A EA+A AYL+ + ++ D D YG + V
Sbjct 124 LGIPWVQAAGEAEAMCAYLNAGGHVDGCLTNDGDTFLYGAQTV 166
> mmu:209334 Gen1, 5830483C08Rik, MGC115970; Gen homolog 1, endonuclease
(Drosophila)
Length=908
Score = 32.7 bits (73), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
LG+ + A EA+A AYL+ + + ++ D D YG + V
Sbjct 124 LGMPWVQAAGEAEAMCAYLNASGHVDGCLTNDGDAFLYGAQTV 166
> cel:T23F6.4 rbd-1; RBD (RNA binding domain) protein family member
(rbd-1); K14787 multiple RNA-binding domain-containing
protein 1
Length=872
Score = 32.3 bits (72), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 35/79 (44%), Gaps = 4/79 (5%)
Query 4 EKRQKRRETARQEALELLKRKQQNLPVDYKE----LMSKCSQSISITPAMVDKVIAACRE 59
E R K ET R++ LE + +NLP KE + K +S ++DK AC+
Sbjct 262 ESRDKEEETVREKILETGRLFLRNLPYATKEDDLQFLFKKYGEVSEVQVVIDKKTGACKG 321
Query 60 LGIRCIVAPFEADAQLAYL 78
I V P A A + L
Sbjct 322 FAIVEFVFPEAAVAAYSAL 340
> dre:386707 fen1, cb879; flap structure-specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=330
Score = 32.3 bits (72), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGC 99
+G+ I AP EA+A A L + ++++ +ED D L +G
Sbjct 148 MGVPYIEAPCEAEASCAALVKAGKVYATATEDMDGLTFGT 187
> dre:100332576 Gen homolog 1, endonuclease-like
Length=179
Score = 32.3 bits (72), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 60 LGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSDLLAYGCKRV 102
LG+ + A EA+A A+L + I+ D D YG + V
Sbjct 63 LGVPWVTAAGEAEAMCAFLDSQGLVDGCITNDGDAFLYGARTV 105
> tpv:TP04_0165 hypothetical protein
Length=708
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 36 MSKCSQSI--SITPAMVDKVIAACRELGIRCIVAPFEADAQLAYLSRTNQIHSAISEDSD 93
+SK SQ I + ++ +++ + G I AP+ A +QL Y + I + + S
Sbjct 90 LSKFSQHIYKEYSEDILQLLMSYLKGKGYEIIRAPYLASSQLVYFMQEGLIDAVVGPPST 149
Query 94 LLAYGCKRVMLKMD 107
LL +G RV++ +D
Sbjct 150 LL-FGVPRVIMGLD 162
Lambda K H
0.318 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4157683456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40