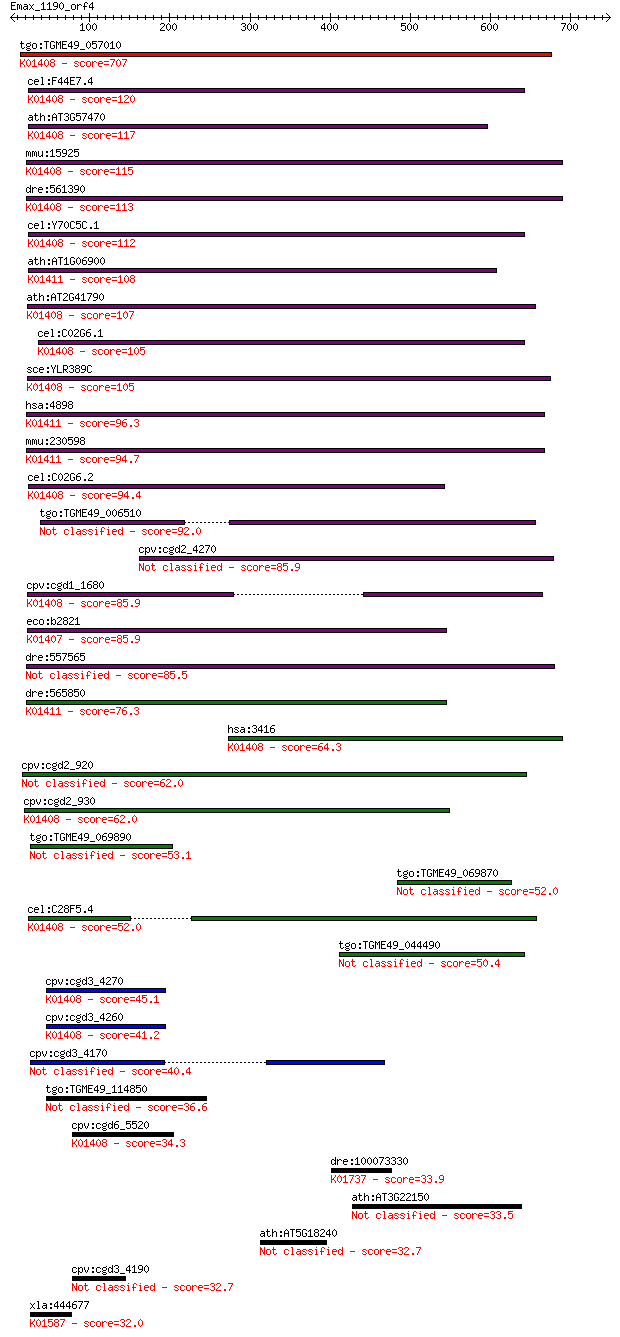
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1190_orf4
Length=749
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 707 0.0
cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 120 2e-26
ath:AT3G57470 peptidase M16 family protein / insulinase family... 117 2e-25
mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI50753... 115 9e-25
dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzym... 113 2e-24
cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24... 112 4e-24
ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptid... 108 6e-23
ath:AT2G41790 peptidase M16 family protein / insulinase family... 107 2e-22
cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 105 9e-22
sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1... 105 9e-22
hsa:4898 NRD1, hNRD1, hNRD2; nardilysin (N-arginine dibasic co... 96.3 5e-19
mmu:230598 Nrd1, 2600011I06Rik, AI875733, MGC25477, NRD-C; nar... 94.7 1e-18
cel:C02G6.2 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 94.4 2e-18
tgo:TGME49_006510 peptidase M16 domain containing protein (EC:... 92.0 7e-18
cpv:cgd2_4270 secreted insulinase-like peptidase 85.9 5e-16
cpv:cgd1_1680 insulinase like protease, signal peptide ; K0140... 85.9 5e-16
eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.... 85.9 6e-16
dre:557565 nrd1, si:dkey-171o17.4; nardilysin (N-arginine diba... 85.5 8e-16
dre:565850 fk24c07; wu:fk24c07; K01411 nardilysin [EC:3.4.24.61] 76.3 4e-13
hsa:3416 IDE, FLJ35968, INSULYSIN; insulin-degrading enzyme (E... 64.3 2e-09
cpv:cgd2_920 peptidase'insulinase-like peptidase' 62.0 8e-09
cpv:cgd2_930 peptidase'insulinase-like peptidase' ; K01408 ins... 62.0 9e-09
tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56) 53.1 4e-06
tgo:TGME49_069870 hypothetical protein 52.0 8e-06
cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 52.0 1e-05
tgo:TGME49_044490 insulin-degrading enzyme, putative (EC:3.4.2... 50.4 2e-05
cpv:cgd3_4270 peptidase'insulinase like peptidase' ; K01408 in... 45.1 0.001
cpv:cgd3_4260 peptidase'insulinase like peptidase' ; K01408 in... 41.2 0.014
cpv:cgd3_4170 secreted insulinase like peptidase 40.4 0.028
tgo:TGME49_114850 hypothetical protein 36.6 0.38
cpv:cgd6_5520 peptidase'insulinase like peptidase' ; K01408 in... 34.3 1.8
dre:100073330 pts, zgc:162276; 6-pyruvoyltetrahydropterin synt... 33.9 2.6
ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein 33.5 3.3
ath:AT5G18240 MYR1; MYR1 (MYb-related protein 1); transcriptio... 32.7 5.0
cpv:cgd3_4190 secreted insulinase like peptidase 32.7 6.1
xla:444677 MGC84289 protein; K01587 phosphoribosylaminoimidazo... 32.0 9.7
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 707 bits (1825), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 328/664 (49%), Positives = 470/664 (70%), Gaps = 3/664 (0%)
Query 14 QSFSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVD 73
QS E SLWVAF LP T+TSYKKQPT +LTYL EY+G+GSL+KRLR +GLAD +S D
Sbjct 281 QSVGGESSLWVAFSLPPTITSYKKQPTGILTYLFEYSGDGSLSKRLRTMGLADEVSVVAD 340
Query 74 RNSISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTT 133
R S+STL +KVDL KGA+ RG VL+E+FSYIN L++ GV + +S++++QS +DFHT+
Sbjct 341 RTSVSTLFAVKVDLASKGASERGAVLEEVFSYINLLKNEGVDSKTISSISEQSLVDFHTS 400
Query 134 QPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPD 193
QP M+E ARLAHNLLTYEPYHV+AGDSLL+D D + NQLL +M+ AIIAF+DP
Sbjct 401 QPDPPAMNEVARLAHNLLTYEPYHVLAGDSLLVDPDAQFVNQLLDKMTSDHAIIAFADPQ 460
Query 194 FTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILP 253
F DSF+ +P+YG+++++ +LP+ + +T SP A+++PP+L H+P+ +L +LP
Sbjct 461 FKRNNDSFDVEPFYGIEYKITNLPKEQRRRLETVTPSPGAYKIPPALKHVPRPEDLHLLP 520
Query 254 GLLGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLAL 313
L G++ PEL+ + + G AVWWQGQG +PR+ + + + ++ SRTQ +L +
Sbjct 521 ALGGMSIPELLGDSNTSGGHAVWWQGQGTLPVPRVHANIKARTQRSRTNMASRTQATLLM 580
Query 314 AAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMVE 373
AA+AE L EETVD + CG++HS+ G G + F AYT QL ++M VA + DP VE
Sbjct 581 AALAEQLDEETVDLKQCGISHSVGVSGDGLLLAFAAYTPKQLRQVMAVVASKIQDP-QVE 639
Query 374 PERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQ--QTNYDDSF 431
+RF+RIKQ+ ++ + D A+ +A+EHA+ AA++L RNDA SRKDLL L+ T+ +++
Sbjct 640 QDRFDRIKQRMIEELEDSASQVAYEHAIAAASVLLRNDANSRKDLLRLLKSSSTSLNETL 699
Query 432 AKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEAT 491
+LK VH DAF+MGNID+ + ++V+ FL+ +GFT I DA SL ++Q+ IEA
Sbjct 700 KTFRDLKAVHADAFIMGNIDKADANSVVQSFLQDSGFTQIPMKDAAQSLVVDQRAPIEAL 759
Query 492 LANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKES 551
+ANPI D +HA++VQ+QLG+PSIE+RVNLAVL Q LNRR++D LRTE QLGYI GA+
Sbjct 760 IANPIPKDVNHATVVQYQLGVPSIEERVNLAVLGQMLNRRLFDRLRTEEQLGYIVGARSY 819
Query 552 QAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKME 611
+S L+C +EG++ HPDE+ +ID+EL K ++L ++ + E+ WKE+A A+L K
Sbjct 820 IDSSVESLRCVLEGSRKHPDEIADLIDKELWKMNDHLQSISDGELDHWKESARAELEKPT 879
Query 612 ANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKLSDPSRRMVVKLIA 671
F E+F +S +I H +CF KRDLE+ YL+ +F+RKQL RT+ KL +P+RR+V ++
Sbjct 880 ETFYEEFGRSWGQIANHGHCFNKRDLELIYLNTEFNRKQLSRTYTKLLEPTRRLVGFVLN 939
Query 672 DLEP 675
+ P
Sbjct 940 HVTP 943
> cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=1051
Score = 120 bits (301), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 138/634 (21%), Positives = 266/634 (41%), Gaps = 31/634 (4%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGI 83
++F P + QP +++L+ + G GSL L+ LG + D ++ + G+
Sbjct 337 ISFPFPDLNGEFLSQPGHYISHLIGHEGPGSLLSELKRLGWVSSLQS--DSHTQAAGFGV 394
Query 84 ---KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQPSS 137
+DL+ +G H ++Q +F+YI L+ G + +A+ S + F QP +
Sbjct 395 YNVTMDLSTEGLEHVDEIIQLMFNYIGMLQSAGPKQWVHDELAELSAVKFRFKDKEQPMT 454
Query 138 SIMDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTS 196
++ AA L Y P+ H+++ LL +P +LL +SP+ + F
Sbjct 455 MAINVAASLQ-----YIPFEHILSSRYLLTKYEPERIKELLSMLSPANMQVRVVSQKFKG 509
Query 197 KVDSFETDPYYGVQFRVLDL-PQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
+ + +P YG + +V D+ P+ L S +A +P +I + K +
Sbjct 510 Q-EGNTNEPVYGTEMKVTDISPETMKKYENALKTSHHALHLPEKNEYIATNFDQKPRESV 568
Query 256 LGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAA 315
P LIS+ G + VW++ + +P+ +L + + SL L
Sbjct 569 KN-EHPRLISDDGW---SRVWFKQDDEYNMPKQETKLALTTPMVAQNPRMSLLSSLWLWC 624
Query 316 IAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMVEPE 375
+++ L EET + G+ L G M Y E Q + +H+A +++ ++
Sbjct 625 LSDTLAEETYNADLAGLKCQLESSPFGVQMRVYGYDEKQ-ALFAKHLANRMTN-FKIDKT 682
Query 376 RFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD--SFAK 433
RF+ + + + + + A S + +L + +S++ LL +D FAK
Sbjct 683 RFDVLFESLKRALTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLEDVQGFAK 742
Query 434 LNELKNVHVDAFVMGNIDRDQSLTMVEEFLE-----QAGFTPIDHDDAVASLAMEQKQTI 488
L+ H++ FV GN +++ + +E ++ P+ ++ ++
Sbjct 743 -EMLQAFHMELFVHGNSTEKEAIQLSKELMDVLKSAAPNSRPLYRNEHNPRRELQLNNGD 801
Query 489 EATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGA 548
E + K V +Q+G+ + D + ++ Q + +++LRT LGYI
Sbjct 802 EYVYRHLQKTHDVGCVEVTYQIGVQNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWT 861
Query 549 KESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLT 608
T L V+G K+ D V++ I+ L ++ +A MP+ E A+L
Sbjct 862 GSRLNCGTVALNVIVQGPKS-VDHVLERIEVFLESVRKEIAEMPQEEFDNQVSGMIARLE 920
Query 609 KMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYL 642
+ S F++ EI F +R+ EV L
Sbjct 921 EKPKTLSSRFRRFWNEIECRQYNFARREEEVALL 954
> ath:AT3G57470 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=891
Score = 117 bits (292), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 125/597 (20%), Positives = 243/597 (40%), Gaps = 49/597 (8%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPA-VDRNSISTLLG 82
V++ + +++ Y++ P L L+ + GEGSL L++LG A G+ D + +
Sbjct 203 VSWPVTPSISHYEEAPCRYLGDLIGHEGEGSLFHALKILGWATGLYAGEADWSMEYSFFN 262
Query 83 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDE 142
+ +DLT G H +L +F YI L+ GV + ++ +FH Q +
Sbjct 263 VSIDLTDAGHEHMQDILGLLFEYIKVLQQSGVSQWIFDELSAICEAEFH-YQAKIDPISY 321
Query 143 AARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFE 202
A ++ N+ Y H + G SL +P + ++L E+SP+ I + F + D E
Sbjct 322 AVDISSNMKIYPTKHWLVGSSLPSKFNPAIVQKVLDELSPNNVRIFWESNKFEGQTDKVE 381
Query 203 TDPYYGVQFRVLDLPQ-------HHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
P+Y + + + + A + +L +PN F P+ + + I P L
Sbjct 382 --PWYNTAYSLEKITKFTIQEWMQSAPDVNLLLPTPNVFI--PTDFSLKDLKDKDIFPVL 437
Query 256 LGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAA 315
L T + + +W++ F P+ V+++ + + + + +
Sbjct 438 LRKT-----------SYSRLWYKPDTKFFKPKAYVKMDFNCPLAVSSPDAAVLSDIFVWL 486
Query 316 IAEHLQEETVDFQNCGVTHSLAFKGTGFHM---GFEAYTEGQLSKLMEHVAKLLSDPSMV 372
+ ++L E Q G+ + L+ GF + GF L +++ +AK V
Sbjct 487 LVDYLNEYAYYAQAAGLDYGLSLSDNGFELSLAGFNHKLRILLEAVIQKIAKF-----EV 541
Query 373 EPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD--S 430
+P+RF IK+ K + E A +++ ++ + + L+AL +D +
Sbjct 542 KPDRFSVIKETVTKAYQNNKFQQPHEQATNYCSLVLQDQIWPWTEELDALSHLEAEDLAN 601
Query 431 FAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFT---PIDH--------DDAVAS 479
F + L V+ ++ GN+++D++ +MV+ +E FT PI + V
Sbjct 602 FVPM-LLSRTFVECYIAGNVEKDEAESMVKH-IEDVLFTDSKPICRPLFPSQFLTNRVTE 659
Query 480 LAMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTE 539
L K +N D++ A + Q+ L + + + LRT
Sbjct 660 LGTGMKHFYYQEGSN--SSDENSALVHYIQVHKDEFSMNSKLQLFELIAKQDTFHQLRTI 717
Query 540 AQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEM 596
QLGYI S + +Q ++ + P + ++ L + NM + E
Sbjct 718 EQLGYITSLSLSNDSGVYGVQFIIQSSVKGPGHIDSRVESLLKDLESKFYNMSDEEF 774
> mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI507533;
insulin degrading enzyme (EC:3.4.24.56); K01408 insulysin
[EC:3.4.24.56]
Length=1019
Score = 115 bits (287), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 147/690 (21%), Positives = 282/690 (40%), Gaps = 46/690 (6%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTL 80
+L+V F +P YK P L +L+ + G GSL L+ G + + + +
Sbjct 312 NLYVTFPIPDLQQYYKSNPGHYLGHLIGHEGPGSLLSELKSKGWVNTLVGGQKEGARGFM 371
Query 81 LGI-KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSI 139
I VDLT++G H ++ +F YI LR G + + + F
Sbjct 372 FFIINVDLTEEGLLHVEDIILHMFQYIQKLRAEGPQEWVFQECKDLNAVAFRFKDKERP- 430
Query 140 MDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 199
+++A L Y V+ + LL + P L + +L ++ P +A F K D
Sbjct 431 RGYTSKIAGKLHYYPLNGVLTAEYLLEEFRPDLIDMVLDKLRPENVRVAIVSKSFEGKTD 490
Query 200 SFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNA-FRMPPSLMHIPKASELKILPGLLGL 258
T+ +YG Q++ +P+ + A N F++P IP E+ L
Sbjct 491 --RTEQWYGTQYKQEAIPED--IIQKWQNADLNGKFKLPTKNEFIPTNFEILSLEKD-AT 545
Query 259 TEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIAE 318
P LI + A + +W++ F +P+ + D L L L + +
Sbjct 546 PYPALIKD---TAMSKLWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAYLYLELLKD 602
Query 319 HLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQ---LSKLMEHVAKLLSDPSMVEPE 375
L E + G+++ L G ++ + Y + Q L K+ E +A ++ +
Sbjct 603 SLNEYAYAAELAGLSYDLQNTIYGMYLSVKGYNDKQPILLKKITEKMATF-----EIDKK 657
Query 376 RFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAKLN 435
RFE IK+ M+ + + +HA+ +L A+++ +L AL A +
Sbjct 658 RFEIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIP 717
Query 436 EL-KNVHVDAFVMGNIDRDQSL---TMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEAT 491
+L +H++A + GNI + +L MVE+ L I+H L + + E
Sbjct 718 QLLSRLHIEALLHGNITKQAALGVMQMVEDTL-------IEHAHTKPLLPSQLVRYREVQ 770
Query 492 LANPIKG-------DKDHASL---VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQ 541
L P +G ++ H + + +Q + S + + L + Q ++ +++LRT+ Q
Sbjct 771 L--PDRGWFVYQQRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQ 828
Query 542 LGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKE 601
LGYI + +A L+ ++ K P + ++ L ++ + +M E + +
Sbjct 829 LGYIVFSGPRRANGIQGLRFIIQSEKP-PHYLESRVEAFLITMEKAIEDMTEEAFQKHIQ 887
Query 602 AAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKL--S 659
A + S + K EI + + + ++EV YL ++ ++R + ++
Sbjct 888 ALAIRRLDKPKKLSAECAKYWGEIISQQYNYDRDNIEVAYLKT-LTKDDIIRFYQEMLAV 946
Query 660 DPSRRMVVKLIADLEPEKEVTLIGEAKAQD 689
D RR V + ++GE +Q+
Sbjct 947 DAPRRHKVSVHVLAREMDSCPVVGEFPSQN 976
> dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=978
Score = 113 bits (283), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 142/689 (20%), Positives = 278/689 (40%), Gaps = 44/689 (6%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTL 80
+L+V F +P YK P L +L+ + G GSL L+ G + + + +
Sbjct 271 NLYVTFPIPDLQKYYKSNPGHYLGHLIGHEGPGSLLSELKSKGWVNTLVGGQKEGARGFM 330
Query 81 LGI-KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQPS 136
I VDLT++G H ++ +F YI LR G + + + F +P
Sbjct 331 FFIINVDLTEEGLLHVEDIIFHMFQYIQKLRTEGPQEWVFQECKDLNTVAFRFKDKERPR 390
Query 137 SSIMDEAARLAHNLLTYEPYH-VVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
A LL Y P ++A + LL + P L +L ++ P +A F
Sbjct 391 GYTSKVAG-----LLHYYPLEEILAAEYLLEEFRPDLIEMVLDKLRPENVRVAVVSKSFE 445
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNA-----FRMPPSLMHIPKASELK 250
+ D T+ +YG Q++ A+ + NA F++P IP E+
Sbjct 446 GQTD--RTEEWYGTQYK------QEAITDEAIKKWDNADLNGKFKLPMKNEFIPTNFEIY 497
Query 251 ILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGS 310
L + P LI + A + VW++ F +P+ + D L
Sbjct 498 PLEKD-SPSAPTLIKD---TAMSKVWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAY 553
Query 311 LALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQ---LSKLMEHVAKLLS 367
L L + + L E + G+++ L G ++ + Y + Q L K++E +A
Sbjct 554 LYLELLKDSLNEYAYAAELAGLSYDLQNTVYGMYLSVKGYNDKQHILLKKIIEKMATF-- 611
Query 368 DPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNY 427
++ +RF+ IK+ M+ + + +HA+ +L A+++ +L +AL
Sbjct 612 ---EIDEKRFDIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDELRDALDDVTL 668
Query 428 DDSFAKLNEL-KNVHVDAFVMGNIDRDQSLTMVE----EFLEQAGFTPIDHDDAVASLAM 482
A + +L +H++A + GNI + +L M++ +E A P+ + +
Sbjct 669 PRLKAFIPQLLSRLHIEALLHGNITKQSALEMMQMLEDTLIEHAHTKPLLPSQLIRYREV 728
Query 483 EQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQL 542
+ + + + +Q + + + + L + Q ++ +++LRT+ QL
Sbjct 729 QVPDGGWYVYQQRNEVHNNCGIEIYYQTDMQNTHENMLLELFCQIISEPCFNTLRTKEQL 788
Query 543 GYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEA 602
GYI + +A L+ ++ KA P + ++ L ++ + M + + +A
Sbjct 789 GYIVFSGPRRANGVQGLRFIIQSEKA-PHYLESRVEAFLKTMEKSVEEMGDEAFQKHIQA 847
Query 603 AHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKL--SD 660
+ + + K EI + F + ++EV YL +++ +++ + L D
Sbjct 848 LAIRRLDKPKKLAAECAKYWGEIISQQYNFDRDNIEVAYLKT-LTKEHIMQFYRDLLAID 906
Query 661 PSRRMVVKLIADLEPEKEVTLIGEAKAQD 689
RR V + L+GE AQ+
Sbjct 907 APRRHKVSVHVLSREMDSCPLVGEFPAQN 935
> cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=985
Score = 112 bits (281), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 136/637 (21%), Positives = 268/637 (42%), Gaps = 37/637 (5%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGI-SPAVDRNSISTLLG 82
++F P + QP +++L+ + G GSL L+ LG + S V + +
Sbjct 278 ISFPFPDLTGEFLSQPEHYISHLIGHEGHGSLLSELKRLGWVVSLQSGYVVQAAGFGNFQ 337
Query 83 IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQPSSSI 139
+ ++L+ +G H ++Q +F+YI ++ G + +A+ + F QP +
Sbjct 338 VGIELSTEGLEHVDEIIQLMFNYIGMMQSSGPKQWVHEELAELRAVTFRFKDKEQPMAMA 397
Query 140 MDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKV 198
AARL P+ HV++ LL + +P +LL + PS I F +
Sbjct 398 SCVAARLQRI-----PFKHVLSSPHLLTNYEPVRIKELLSMLIPSNMKIQVVSQKFKGQ- 451
Query 199 DSFETDPYYGVQFRVLDLPQHHAVAMA-VLTASPNAFRMPPSLMHIPKASELKILPG-LL 256
+ +P YG + +V + L S +A +P +I A++ P L+
Sbjct 452 EGNTNEPVYGTEIKVTRISSETMQKYEEALKTSHHALHLPEKNQYI--ATKFDQKPRELV 509
Query 257 GLTEPELISEQGGNAGTAVWWQGQGAFAIPR--ITVQLNGSILKD--KADLLSRTQGSLA 312
P LI++ + + VW++ + +P+ + L I+ + LLSR L
Sbjct 510 KSDHPRLIND---DEWSRVWFKQDDEYKMPKQETKLALTTPIVSQSPRMTLLSR----LW 562
Query 313 LAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMV 372
L +++ L EE+ + G+ + L G M Y E Q + +H+ K L + +
Sbjct 563 LRCLSDSLAEESYSAKVAGLNYELESSFFGVQMRVSGYAEKQ-ALFSKHLTKRLFN-FKI 620
Query 373 EPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD--S 430
+ RF+ + + + + A S + + +L + +S++ LL + +D
Sbjct 621 DQTRFDVLFDSLKRDLTNHAFSQPYVLSQHYTELLVVDKEWSKQQLLAVCESVKLEDVQR 680
Query 431 FAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLE-----QAGFTPIDHDDAVASLAMEQK 485
F K L+ H++ V GN +++ + ++ ++ P+ ++ + ++
Sbjct 681 FGK-EMLQAFHLELLVYGNSTEKETIQLSKDLIDILKSAAPSSRPLFRNEHILRREIQLN 739
Query 486 QTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYI 545
E + V +Q+G+ + D + ++ + +D+LRT+ LGYI
Sbjct 740 NGDEYIYRHLQTTHDVGCVQVTYQIGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYI 799
Query 546 AGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHA 605
+ T LQ V+G K+ D V++ I+ L ++ + MP+ E A
Sbjct 800 VWTRTHFNCGTVALQILVQGPKS-VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIA 858
Query 606 KLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYL 642
+L + S FKK +EI FT+ + +V+ L
Sbjct 859 QLEEKPKTLSCRFKKFWDEIECRQYNFTRIEEDVELL 895
> ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01411 nardilysin [EC:3.4.24.61]
Length=1024
Score = 108 bits (271), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 129/610 (21%), Positives = 247/610 (40%), Gaps = 47/610 (7%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAV-----DRNSIS 78
+ + LP ++Y K+P L +LL + G GSL L+ G A +S V +R+S++
Sbjct 334 LTWTLPPLRSAYVKKPEDYLAHLLGHEGRGSLHSFLKAKGWATSLSAGVGDDGINRSSLA 393
Query 79 TLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTT--QPS 136
+ G+ + LT G ++ I+ Y+ LRD + + ++DF QP+
Sbjct 394 YVFGMSIHLTDSGLEKIYDIIGYIYQYLKLLRDVSPQEWIFKELQDIGNMDFRFAEEQPA 453
Query 137 SSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTS 196
D AA L+ N+L Y HV+ GD + DP+L L+ +P I S
Sbjct 454 D---DYAAELSENMLAYPVEHVIYGDYVYQTWDPKLIEDLMGFFTPQNMRIDVVSKSIKS 510
Query 197 KVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLL 256
+ F+ +P++G + D+P + + + N+ +P IP ++ + +
Sbjct 511 --EEFQQEPWFGSSYIEEDVPLSLMESWSNPSEVDNSLHLPSKNQFIPCDFSIRAINSDV 568
Query 257 ---GLTEPELISEQGGNAGTAVWWQGQGAFAIPR----ITVQLNGSILKDKADLLSRTQG 309
+ P I ++ W++ F +PR + L G+ K LL+
Sbjct 569 DPKSQSPPRCIIDE---PFMKFWYKLDETFKVPRANTYFRINLKGAYASVKNCLLTE--- 622
Query 310 SLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHM---GFEAYTEGQLSKLMEHVAKLL 366
L + + + L E + SL+ G + GF LSK++ +AK
Sbjct 623 -LYINLLKDELNEIIYQASIAKLETSLSMYGDKLELKVYGFNEKIPALLSKILA-IAKSF 680
Query 367 SDPSMVEPERFERIKQKQMKLVADPATSMA-FEHALEAAAILTRNDAFSRKDLLNALQQT 425
M ERF+ IK+ + + T+M H+ L + + L+ L
Sbjct 681 ----MPNLERFKVIKENMERGFRN--TNMKPLNHSTYLRLQLLCKRIYDSDEKLSVLNDL 734
Query 426 NYDDSFAKLNELKN-VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPI----DHDDAVASL 480
+ DD + + EL++ + ++A GN+ D+++ + F + P+ H + +
Sbjct 735 SLDDLNSFIPELRSQIFIEALCHGNLSEDEAVNISNIFKDSLTVEPLPSKCRHGEQITCF 794
Query 481 AMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQF---LNRRIYDSLR 537
M K + + N K + + + +Q+ + AVL F + +++ LR
Sbjct 795 PMGAKLVRDVNVKN--KSETNSVVELYYQIEPEEAQSTRTKAVLDLFHEIIEEPLFNQLR 852
Query 538 TEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMA 597
T+ QLGY+ V+ +K P ++ +D + + L + +
Sbjct 853 TKEQLGYVVECGPRLTYRVHGFCFCVQSSKYGPVHLLGRVDNFIKDIEGLLEQLDDESYE 912
Query 598 RWKEAAHAKL 607
++ A+L
Sbjct 913 DYRSGMIARL 922
> ath:AT2G41790 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=970
Score = 107 bits (267), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 131/658 (19%), Positives = 268/658 (40%), Gaps = 50/658 (7%)
Query 22 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSIS-TL 80
L V++ + ++ Y + P+ L +L+ + GEGSL L+ LG A G+S ++ +
Sbjct 274 LGVSWPVTPSIHHYDEAPSQYLGHLIGHEGEGSLFHALKTLGWATGLSAGEGEWTLDYSF 333
Query 81 LGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQ---PSS 137
+ +DLT G H +L +F+YI L+ GV + ++ FH P S
Sbjct 334 FKVSIDLTDAGHEHMQEILGLLFNYIQLLQQTGVCQWIFDELSAICETKFHYQDKIPPMS 393
Query 138 SIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSK 197
I+D +A N+ Y + G SL +P + +++ E+SPS I + F +
Sbjct 394 YIVD----IASNMQIYPTKDWLVGSSLPTKFNPAIVQKVVDELSPSNFRIFWESQKFEGQ 449
Query 198 VDSFETDPYYGVQFRVLDLPQ-------HHAVAMAVLTASPNAFRMPPSLMHIPKASELK 250
D + +P+Y + + + A + + +PN F P+ + + A + +
Sbjct 450 TD--KAEPWYNTAYSLEKITSSTIQEWVQSAPDVHLHLPAPNVFI--PTDLSLKDADDKE 505
Query 251 ILPGLLGLTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGS 310
+P LL T + +W++ F+ P+ V+++ + + +
Sbjct 506 TVPVLLRKT-----------PFSRLWYKPDTMFSKPKAYVKMDFNCPLAVSSPDAAVLTD 554
Query 311 LALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPS 370
+ + ++L E Q G+ + ++ GF + Y +L L+E V +++
Sbjct 555 IFTRLLMDYLNEYAYYAQVAGLYYGVSLSDNGFELTLLGYNH-KLRILLETVVGKIANFE 613
Query 371 MVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDS 430
V+P+RF IK+ K + + A+ +++ ++ + + L+ L +D
Sbjct 614 -VKPDRFAVIKETVTKEYQNYKFRQPYHQAMYYCSLILQDQTWPWTEELDVLSHLEAED- 671
Query 431 FAKLNE--LKNVHVDAFVMGNIDRDQSLTMVEEFLEQAGFT----------PIDH-DDAV 477
AK L ++ ++ GN++ +++ +MV+ +E F P H + V
Sbjct 672 VAKFVPMLLSRTFIECYIAGNVENNEAESMVKH-IEDVLFNDPKPICRPLFPSQHLTNRV 730
Query 478 ASLAMEQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLR 537
L K +NP D++ A + Q+ + L + + + LR
Sbjct 731 VKLGEGMKYFYHQDGSNP--SDENSALVHYIQVHRDDFSMNIKLQLFGLVAKQATFHQLR 788
Query 538 TEAQLGYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMA 597
T QLGYI + + +Q ++ + P + ++ L + L M +
Sbjct 789 TVEQLGYITALAQRNDSGIYGVQFIIQSSVKGPGHIDSRVESLLKNFESKLYEMSNEDFK 848
Query 598 RWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTF 655
A + N E+ + EI + + F +++ EV L +++L+ F
Sbjct 849 SNVTALIDMKLEKHKNLKEESRFYWREIQSGTLKFNRKEAEVSAL-KQLQKQELIDFF 905
> cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=980
Score = 105 bits (261), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 137/640 (21%), Positives = 261/640 (40%), Gaps = 51/640 (7%)
Query 36 KKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSIST---LLGIKVDLTQKGA 92
K+ +L+ + G GSL L+ LG + + D N+I+ +L + +DL+ G
Sbjct 263 KRIDRKFFAHLIRHKGPGSLLVELKRLGWVNSLKS--DSNTIAAGFGILNVTMDLSTGGL 320
Query 93 AHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFH---TTQPSSSIMDEAARLAHN 149
+ ++Q + +YI L+ G + +A S + F QP ++ AA L
Sbjct 321 ENVDEIIQLMLNYIGMLKSFGPQQWIHDELADLSDVKFRFKDKEQPMKMAINIAASLQ-- 378
Query 150 LLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYG 208
Y P H+++ LL +P +LL ++PS ++ F + + +P YG
Sbjct 379 ---YIPIEHILSSRYLLTKYEPERIKELLSTLTPSNMLVRVVSQKFKEQ-EGNTNEPVYG 434
Query 209 VQFRVLDL-PQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLGLTE-PELISE 266
+ +V D+ P+ L S +A +P +I A+ P E P+LIS+
Sbjct 435 TEMKVTDISPEKMKKYENALKTSHHALHLPEKNEYI--ATNFGQKPRESVKNEHPKLISD 492
Query 267 QGGNAGTAVWW---------QGQGAFAI--------PRITVQLNGSILKDKADLLSRTQG 309
G + VW+ + + FA+ PRI++ ++ L D+LS
Sbjct 493 DG---WSRVWFKQDDEYNMPKQETKFALTTPIVSQNPRISL-ISSLWLWCFCDILSEETY 548
Query 310 SLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDP 369
+ ALA + + Q A + + Y E Q ++H+ + +
Sbjct 549 NAALAGLGCQFELSPFGVQKQSTDGREAERHASLTLHVYGYDEKQ-PLFVKHLTSCMIN- 606
Query 370 SMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD 429
++ RFE + + + + + A S + +L + +S++ LL ++
Sbjct 607 FKIDRTRFEVLFESLKRTLTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLEN 666
Query 430 --SFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLE-----QAGFTPIDHDDAVASLAM 482
FA+ L+ H++ FV GN +++ + +E ++ P+ ++
Sbjct 667 VQGFAR-EMLQAFHMELFVHGNSTEKEAIQLSKELMDILKSAAPNSRPLYRNEHNPRREF 725
Query 483 EQKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQL 542
+ E + K V +Q+G+ + D + ++ Q + ++D+LRT L
Sbjct 726 QLNNGDEYIYRHLQKTHDAGCVEVTYQIGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEAL 785
Query 543 GYIAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEA 602
GYI L FV+G K+ D V++ I+ L ++ + MP+ E +
Sbjct 786 GYIVWTGCRFNCGAVALNIFVQGPKS-VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAG 844
Query 603 AHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYL 642
A+L + S FK+ +I F +R+ EVK L
Sbjct 845 MIARLEEKPKTLSNRFKRFWYQIECRQYDFARREKEVKVL 884
> sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1p,
in N-terminal processing of pro-A-factor to the mature
form; member of the insulin-degrading enzyme family (EC:3.4.24.-);
K01408 insulysin [EC:3.4.24.56]
Length=1027
Score = 105 bits (261), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 132/668 (19%), Positives = 286/668 (42%), Gaps = 35/668 (5%)
Query 22 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSI-STL 80
L ++F +P ++ +P +L++L+ + G GSL L+ LG A+ +S S +
Sbjct 323 LEISFTVPDMEEHWESKPPRILSHLIGHEGSGSLLAHLKKLGWANELSAGGHTVSKGNAF 382
Query 81 LGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQ---PSS 137
+ +DLT G H V+ IF YI L++ + + + S+ F Q PSS
Sbjct 383 FAVDIDLTDNGLTHYRDVIVLIFQYIEMLKNSLPQKWIFNELQDISNATFKFKQAGSPSS 442
Query 138 SIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSK 197
++ A L + Y P + LL +P L Q + P + + +
Sbjct 443 TVSSLAKCLEKD---YIPVSRILAMGLLTKYEPDLLTQYTDALVPENSRVTL----ISRS 495
Query 198 VDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLG 257
+++ + +YG ++V+D P M +P A +P + ++ + G+
Sbjct 496 LETDSAEKWYGTAYKVVDYPADLIKNMKSPGLNP-ALTLPRPNEFVSTNFKVDKIDGIKP 554
Query 258 LTEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIA 317
L EP L+ + + +W++ F PR + L+ + A +++ +L
Sbjct 555 LDEPVLLL---SDDVSKLWYKKDDRFWQPRGYIYLSFKLPHTHASIINSMLSTLYTQLAN 611
Query 318 EHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMVEP--E 375
+ L++ V + ++F T + G G KL+ + + L + EP +
Sbjct 612 DALKD--VQYDAACADLRISFNKT--NQGLAITASGFNEKLIILLTRFLQGVNSFEPKKD 667
Query 376 RFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAKLN 435
RFE +K K ++ + + + + + ++S + L ++ ++ +
Sbjct 668 RFEILKDKTIRHLKNLLYEVPYSQMSNYYNAIINERSWSTAEKLQVFEKLTFEQLINFIP 727
Query 436 EL-KNVHVDAFVMGNIDRDQSL---TMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEAT 491
+ + V+ + + GNI +++L ++++ + ++ + S + + +T
Sbjct 728 TIYEGVYFETLIHGNIKHEEALEVDSLIKSLIPNNIHNLQVSNNRLRSYLLPKGKTFRYE 787
Query 492 LANPIKGDKDHASLVQF--QLGIPSIEDRVNLA-VLTQFLNRRIYDSLRTEAQLGYIAGA 548
A +K ++ S +Q QL + S ED L+ + Q ++ +D+LRT+ QLGY+ +
Sbjct 788 TA--LKDSQNVNSCIQHVTQLDVYS-EDLSALSGLFAQLIHEPCFDTLRTKEQLGYVVFS 844
Query 549 KESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLT 608
TA ++ ++ P + I+ + L +MPE + + KEA L
Sbjct 845 SSLNNHGTANIRILIQSEHTTP-YLEWRINNFYETFGQVLRDMPEEDFEKHKEALCNSLL 903
Query 609 KMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAK--LSDPSRRMV 666
+ N +E+ + I+ FT R + K + N +++Q++ + +S+ + +++
Sbjct 904 QKFKNMAEESARYTAAIYLGDYNFTHRQKKAKLVAN-ITKQQMIDFYENYIMSENASKLI 962
Query 667 VKLIADLE 674
+ L + +E
Sbjct 963 LHLKSQVE 970
> hsa:4898 NRD1, hNRD1, hNRD2; nardilysin (N-arginine dibasic
convertase) (EC:3.4.24.61); K01411 nardilysin [EC:3.4.24.61]
Length=1151
Score = 96.3 bits (238), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 141/675 (20%), Positives = 280/675 (41%), Gaps = 59/675 (8%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGS----LAKRLRLLGLADGIS-PAVDRN 75
+L + + LP Y+ +P +++L+ + G+GS L K+ L L G ++N
Sbjct 439 ALTITWALPPQQQHYRVKPLHYISWLVGHEGKGSILSFLRKKCWALALFGGNGETGFEQN 498
Query 76 SISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQP 135
S ++ I + LT +G H V +F Y+ L+ G + + + +FH Q
Sbjct 499 STYSVFSISITLTDEGYEHFYEVAYTVFQYLKMLQKLGPEKRIFEEIRKIEDNEFHY-QE 557
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
+ ++ + N+ Y ++ GD LL + P + + L ++ P KA +
Sbjct 558 QTDPVEYVENMCENMQLYPLQDILTGDQLLFEYKPEVIGEALNQLVPQKANLVLLSGANE 617
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
K D E ++G Q+ + D+ + A L S F + P L H+P +E K +
Sbjct 618 GKCDLKEK--WFGTQYSIEDIEN----SWAELWNS--NFELNPDL-HLP--AENKYIATD 666
Query 256 LGLT-----EPELISEQGGNAGTAVWWQGQGAFAIPRITVQ---LNGSILKDKADL-LSR 306
L E E + +W++ F IP+ ++ ++ I K A++ L
Sbjct 667 FTLKAFDCPETEYPVKIVNTPQGCLWYKKDNKFKIPKAYIRFHLISPLIQKSAANVVLFD 726
Query 307 TQGSLALAAIAEHLQEETV---DFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVA 363
++ +AE E V +++ H L + GF+ +++++A
Sbjct 727 IFVNILTHNLAEPAYEADVAQLEYKLVAGEHGLIIRVKGFNHKLPLL----FQLIIDYLA 782
Query 364 KLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQ 423
+ S P++ E++K+ ++ P T A + ++ +S D AL
Sbjct 783 EFNSTPAVFTMIT-EQLKKTYFNILIKPETL-----AKDVRLLILEYARWSMIDKYQALM 836
Query 424 QT-NYDDSFAKLNELKN-VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLA 481
+ + + + E K+ + V+ V GN+ +S+ ++ +++ F P++ + V
Sbjct 837 DGLSLESLLSFVKEFKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFKPLEQEMPV---- 892
Query 482 MEQKQTIEATLANPI-------KGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYD 534
Q Q +E + + KGD + V +Q G S+ + + +L + +D
Sbjct 893 --QFQVVELPSGHHLCKVKALNKGDANSEVTVYYQSGTRSLREYTLMELLVMHMEEPCFD 950
Query 535 SLRTEAQLGY--IAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMP 592
LRT+ LGY + + + + K + + V K I+E LS +E + N+
Sbjct 951 FLRTKQTLGYHVYPTCRNTSGILGFSVTVGTQATKYNSEVVDKKIEEFLSSFEEKIENLT 1010
Query 593 EAEMARWKEAAHAKLTKME-ANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQL 651
E E + A KL + E + E+ ++ E+ F + E++ L + FS+ L
Sbjct 1011 E-EAFNTQVTALIKLKECEDTHLGEEVDRNWNEVVTQQYLFDRLAHEIEALKS-FSKSDL 1068
Query 652 LRTFAKLSDPSRRMV 666
+ F P +M+
Sbjct 1069 VNWFKAHRGPGSKML 1083
> mmu:230598 Nrd1, 2600011I06Rik, AI875733, MGC25477, NRD-C; nardilysin,
N-arginine dibasic convertase, NRD convertase 1 (EC:3.4.24.61);
K01411 nardilysin [EC:3.4.24.61]
Length=1161
Score = 94.7 bits (234), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 145/681 (21%), Positives = 279/681 (40%), Gaps = 71/681 (10%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLR----LLGLADGIS-PAVDRN 75
+L + + LP Y+ +P +++L+ + G+GS+ LR L L G ++N
Sbjct 450 ALTITWALPPQQQHYRVKPLHYISWLVGHEGKGSILSYLRKKCWALALFGGNGETGFEQN 509
Query 76 SISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQP 135
S ++ I + LT +G H V +F Y+ L+ G + + + +FH Q
Sbjct 510 STYSVFSISITLTDEGYEHFYEVAHTVFQYLKMLQKLGPEKRVFEEIQKIEDNEFHY-QE 568
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
+ ++ + N+ Y + GD LL + P + + L ++ P KA +
Sbjct 569 QTDPVEYVENMCENMQLYPRQDFLTGDQLLFEYKPEVIAEALNQLVPQKANLVLLSGANE 628
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
+ D E ++G Q+ + D+ + L S F + P L H+P +E K +
Sbjct 629 GRCDLKEK--WFGTQYSIEDIEN----SWTELWKS--NFDLNPDL-HLP--AENKYIATD 677
Query 256 LGLT-----EPELISEQGGNAGTAVWWQGQGAFAIPRITVQ---LNGSILKDKADL-LSR 306
L E E ++ A +W++ F IP+ ++ ++ I K A++ L
Sbjct 678 FTLKAFDCPETEYPAKIVNTAQGCLWYKKDNKFKIPKAYIRFHLISPLIQKSAANVVLFD 737
Query 307 TQGSLALAAIAEHLQEETV---DFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVA 363
++ +AE E V +++ H L + GF+ +L L + +
Sbjct 738 IFVNILTHNLAEPAYEADVAQLEYKLVAGEHGLIIRVKGFNH--------KLPLLFQLII 789
Query 364 KLLSDPSMVEPERF----ERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLL 419
L++ S P F E++K+ ++ P T A + ++ +S D
Sbjct 790 DYLTEFSST-PAVFTMITEQLKKTYFNILIKPETL-----AKDVRLLILEYSRWSMIDKY 843
Query 420 NALQQTNYDDSFAKLNELKN----VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDD 475
AL DS LN +K+ + V+ V GN+ +S+ ++ +++ F P++ +
Sbjct 844 QALMDGLSLDSL--LNFVKDFKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFAPLEREM 901
Query 476 AVASLAMEQKQTIEATLANPI-------KGDKDHASLVQFQLGIPSIEDRVNLAVLTQFL 528
V Q Q +E + + KGD + V +Q G S+ + + +L +
Sbjct 902 PV------QFQVVELPSGHHLCKVRALNKGDANSEVTVYYQSGTRSLREYTLMELLVMHM 955
Query 529 NRRIYDSLRTEAQLGY--IAGAKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSKAKE 586
+D LRT+ LGY + + + + K + + V K I+E LS +E
Sbjct 956 EEPCFDFLRTKQTLGYHVYPTCRNTSGILGFSVTVGTQATKYNSETVDKKIEEFLSSFEE 1015
Query 587 YLANMPEAEMARWKEAAHAKLTKME-ANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDND 645
+ N+ E + A KL + E + E+ ++ E+ F + E++ L +
Sbjct 1016 KIENLTEDAFNT-QVTALIKLKECEDTHLGEEVDRNWNEVVTQQYLFDRLAHEIEALKS- 1073
Query 646 FSRKQLLRTFAKLSDPSRRMV 666
FS+ L+ F P +M+
Sbjct 1074 FSKSDLVSWFKAHRGPGSKML 1094
> cel:C02G6.2 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=816
Score = 94.4 bits (233), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 111/546 (20%), Positives = 228/546 (41%), Gaps = 38/546 (6%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLG- 82
+ F P Y QP + +L+ + G GS++ L+ LG A + P + +I+ G
Sbjct 278 IIFPFPDLNNEYLSQPGHYIAHLIGHKGPGSISSELKRLGWASSLKP--ESKTIAAGFGY 335
Query 83 --IKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIM 140
+ +DL+ +G H ++Q +F+YI L+ G + +A+ S I+F + +
Sbjct 336 FNVTMDLSTEGLEHVDEIIQLMFNYIGMLQSAGPQQWIHEELAELSAIEFR-FKDREPLT 394
Query 141 DEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 199
A ++A N L Y P+ H+++ LL +P +LL ++PS ++ F + +
Sbjct 395 KNAIKVARN-LQYIPFEHILSSRYLLTKYNPERIKELLSTLTPSNMLVRVVSKKFKEQ-E 452
Query 200 SFETDPYYGVQFRVLDL-PQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLGL 258
+P YG + +V D+ P+ L S +A +P +I + K +
Sbjct 453 GNTNEPVYGTEMKVTDISPEKMKKYENALKTSHHALHLPEKNEYIVTKFDQKPRESVKN- 511
Query 259 TEPELISEQGGNAGTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIAE 318
P LIS+ G + VW++ + +P+ +L + + + SL L + +
Sbjct 512 EHPRLISDDG---WSRVWFKQDDEYNMPKQETKLAFTTPIVAQNPIMSLISSLWLWCLND 568
Query 319 HLQEETVDFQNCGVTHSLAFKGTGFH----------------MGFEAYTEGQLSKLMEHV 362
L EET + G+ L G H + Y E Q ++H+
Sbjct 569 TLTEETYNAAIAGLKFQLESGHNGVHEQAGNWLDPERHASITLHVYGYDEKQ-PLFVKHL 627
Query 363 AKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNAL 422
K +++ ++ RF+ + + + + + A S + + +L +S++ LL
Sbjct 628 TKCMTN-FKIDRTRFDVVFESLKRSLTNHAFSQPYMLSKYFNELLVVEKVWSKEQLLAVC 686
Query 423 QQTNYDDSFAKLNEL-KNVHVDAFVMGNIDRDQSLTMVEEFLE----QAGFTPIDHDDAV 477
+D EL + H++ FV GN +++ + E ++ A + + + +
Sbjct 687 DSATLEDVQGFSKELFQAFHLELFVHGNSTEKKAIQLSNELMDILKSAAPNSRLLYRNEH 746
Query 478 ASLAMEQKQTIEATLANPIKGDKDHASL-VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSL 536
Q + + ++ D + V F+ G+ + D +++Q + + + +L
Sbjct 747 NPRREFQLNNGDEYIYRHLQKTHDAGCVEVTFKFGVQNTYDNALAGLISQLIRQPAFSTL 806
Query 537 RTEAQL 542
RT+ L
Sbjct 807 RTKESL 812
> tgo:TGME49_006510 peptidase M16 domain containing protein (EC:4.1.1.70
3.4.24.13 3.4.24.56 3.4.21.10 3.4.24.35 3.2.1.91)
Length=2435
Score = 92.0 bits (227), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 88/392 (22%), Positives = 179/392 (45%), Gaps = 14/392 (3%)
Query 274 AVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIAEHLQEETVDFQNCGVT 333
+V+W+ F P + + + A + G + E + FQ CGV
Sbjct 960 SVFWKNAEPFNKPIVRGYFKLRVSAEDATAQNTLYGKIFATLAGERARTALASFQGCGVD 1019
Query 334 HSLAFKGTGFHMGFEAYTE---GQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVAD 390
++F + +A++E L++L+E V K D V+ F +I ++D
Sbjct 1020 LLMSFTNGALVLEIQAFSELFAPVLARLIE-VLKESQD--NVKQSDFNKIFNTLKVQLSD 1076
Query 391 PATSMAFEHALEAAAILTRNDAFSRKDLLNALQ--QTNYDDSFAKLNEL--KNVHVDAFV 446
+T FE AL+ A + R + FS+ DL +A+ + ++D L ++ KN +D F+
Sbjct 1077 FSTVTPFELALDVALSVVRRNRFSQLDLRSAVTDASSQFEDFKVFLEKVLTKNA-LDVFI 1135
Query 447 MGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEATLANPIKGDKDHASLV 506
MG+ID +++ + E+F P+ ++ S + IE +NPI D +A +
Sbjct 1136 MGDIDYEEARKLAEDFRAALSKQPLPFSESAGSEILNLADDIEIRFSNPIPEDATNAYVS 1195
Query 507 QFQL-GIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG 565
+ P + + V +++ + ++ +D++RT GY+A A + L V+G
Sbjct 1196 LYVTHPPPDMMEMVVYSLIGEVISSPFFDTIRTHWMDGYVAAAAVREVPPAMTLATIVQG 1255
Query 566 AKAHPDEVVKMIDEELSKAKEYLAN--MPEAEMARWKEAAHAKLTKMEANFSEDFKKSAE 623
++ PDE+ + + L++ +E + + EA + R + + +K + +FS+ F +
Sbjct 1256 SQRKPDELERHVCAFLAEMEENIGSSMTTEAFLERLRWLSSSKFHRSATSFSDYFGEVTS 1315
Query 624 EIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTF 655
+I + + CF + L + S +L+++
Sbjct 1316 QIASRNFCFIREQLARLATEKFLSCPAILKSY 1347
Score = 38.9 bits (89), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 55/214 (25%), Positives = 85/214 (39%), Gaps = 57/214 (26%)
Query 38 QPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGIKVDLTQKGAAHRGL 97
QPT++L YLLEY GE +L RL+ GL S + + +LT +G
Sbjct 639 QPTALLEYLLEYPGEAALLNRLKAQGLIADAEYVDYTTSQKAFVALLFELTDEGEEKFED 698
Query 98 VLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYE--- 154
V+ +Y LR +++ + +DF DE AR+A+ TY+
Sbjct 699 VVSATLAYAEQLR---------TSVTETYILDF---------FDEFARVANRSWTYKDPE 740
Query 155 -------------------PYHVVAGDSLL-IDADPRLTNQLLQE------MSPSKAIIA 188
P V+AG + + D L +L+E + + AI+
Sbjct 741 DAVSAVIAAAEKLAVLPQRPDMVIAGGEFVSLPGDRTLLVDVLKEELESFGRARASAIVV 800
Query 189 FSDPDFTSK-----VDSFETDPYYGVQFRVLDLP 217
P+ T++ V +F YGVQF V LP
Sbjct 801 L--PEDTARGSAEVVHAFRP---YGVQFSVSALP 829
> cpv:cgd2_4270 secreted insulinase-like peptidase
Length=1257
Score = 85.9 bits (211), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 125/562 (22%), Positives = 236/562 (41%), Gaps = 61/562 (10%)
Query 162 DSLLIDADPRLTNQLLQEMSPSK-AIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHH 220
DS L + + ++ + PSK II SD +S +F +PY +
Sbjct 567 DSELSNGQITALKEFVKHLEPSKMKIIKLSDSLNSSSEHNFRFEPY----------KTEY 616
Query 221 AVAMAVLTASPNAFRMPPSLMHIPKASEL-KILPGLLGLT-------------EPELISE 266
+++ L S N + +L K EL +PG L + E E+
Sbjct 617 SISKISLENSENTSKTIEALKSTNKIGELLTCVPGDLSIISFSEDKCPGYKSFEKEIQER 676
Query 267 QGGNA----------GTAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAI 316
+ GN G ++W+G +P I + L + D+ + + SL +A +
Sbjct 677 KIGNELQPCPILEEEGLRIFWKGP-IHTVPTINLTLVQRL--PNKDVSNNVRVSL-IANL 732
Query 317 AEHLQEETVD-----FQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAK-LLSDPS 370
LQ +D F+ CG+ +++ F + ++Y+ ++E ++ L+S+
Sbjct 733 HAQLQNSKMDYILSSFKLCGLEADISYSRGRFVINVQSYS-SNFEDIIEKLSNYLVSESR 791
Query 371 MVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQT--NYD 428
+ FE + + + MA++ A + A + ++ +SR L +LQ+T +D
Sbjct 792 LPTKTEFETALTNLKSEILNLSDLMAYDVATDVAQSVYLSNYYSRLQLRESLQKTEITFD 851
Query 429 DSFAKLNELKNV-HVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLAMEQKQT 487
+ K+ ++ +V + DA ++GNI ++S+ +V + I + +A+ +
Sbjct 852 EYIEKIKDIFSVGYFDALIVGNIGYEKSIKLVSRMVGSLVTKKIPYSNAIHDGILNVSGD 911
Query 488 IEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAG 547
I NPI DK++A + F + D + + + LN YD+LRTE Q GY+A
Sbjct 912 IHIKANNPISSDKNNAVVAHFLTPPVDLIDVSIYSSIGEILNSPFYDTLRTEWQDGYVAF 971
Query 548 AKESQAASTALLQCFVEGAKAHPDEVVKMIDEELSK----AKEYLANMPEAEM---ARWK 600
A L V+ A+ + +V + L K +E L + ++E RW
Sbjct 972 ATTKYETPIISLIGAVQSAEKLSETLVCHMFSALKKVSKDVEEDLKEISKSEFEDKIRWF 1031
Query 601 EAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQL-LRTFAKLS 659
+ K+++ F++ + + + +H CF K L S + + KL
Sbjct 1032 GLSKYSSQKLDS-FTKYIEHFGKLVVSHELCFEKNKLIENATQAFISEPNIYIEKLNKLI 1090
Query 660 DP--SRRMV-VKLIADLEPEKE 678
P SRR+V V+LI + P+ +
Sbjct 1091 KPSSSRRLVIVELIGNKSPDNK 1112
> cpv:cgd1_1680 insulinase like protease, signal peptide ; K01408
insulysin [EC:3.4.24.56]
Length=1033
Score = 85.9 bits (211), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 62/229 (27%), Positives = 111/229 (48%), Gaps = 8/229 (3%)
Query 442 VDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEATLANPIKGDKD 501
+ A++ GNI +++SL ++E+F+ + ++ ++ + + I+ L NP+ D +
Sbjct 761 IIAYLQGNISKNKSLYLIEKFVLNSKILSLNDKYSMKKKIHKLTRPIDIALINPVFEDIN 820
Query 502 HASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQC 561
+ L +Q G+PS E++++L L + IYD+LRT QLGYI A +ST LL
Sbjct 821 NTVLAFYQFGVPSFEEKLHLMALQPIIQGYIYDNLRTNKQLGYIIFANIVPISSTRLLVV 880
Query 562 FVEGAKAHPDEVVKMIDE----ELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSED 617
VEG + E ++ I E S K L NM K A + + +F++
Sbjct 881 GVEGDNNNSVEKIESIIRNTLYEFSTRK--LGNMESHMFEDIKSALIQEAKSIGNSFNQK 938
Query 618 FKKSAEEIFAHSNCFTKRDLE--VKYLDNDFSRKQLLRTFAKLSDPSRR 664
+EI + +L+ + Y++N + + L TF+KL + R
Sbjct 939 LNHYWDEIRYVGDLSESFNLQRAIDYINNKMTIEHLYNTFSKLINSKER 987
Score = 61.2 bits (147), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 57/265 (21%), Positives = 116/265 (43%), Gaps = 19/265 (7%)
Query 22 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLL 81
LW+ + PA L S KQP L+Y+L + SL L+ + + + ++
Sbjct 305 LWLIWSFPARLISPVKQPLIYLSYILNSKQKNSLFWFLQKNNYITNSNSVYENYTFGSIF 364
Query 82 GIKVDLTQKGAAHRGLVLQEIFSYINFLRDHG---VGHELVSTMAQQSHIDFHTTQPSSS 138
+++LT +G ++ ++ I+ YIN L++ ++ + ++ ++ I +T SS
Sbjct 365 IYQLELTSEGLKNQFEIIGLIYKYINKLKESKELLKVYQGIRSLTEREFIT-NTEMLESS 423
Query 139 IMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDP-DFTSK 197
M + + ++ Y + ++GD L+ D D L ++L +SP + SD +F+
Sbjct 424 PMHSTSEICSKMIQYGVHAALSGDILIEDVDENLIYEILNAISPFDTLFLVSDEQEFSGT 483
Query 198 VDSFETDPYYGVQFRVLDLPQHHAVAMAVLTAS---PNAFRMPPSLMHIPKASELKILPG 254
+ F + V+ + D+P A + N ++P P L+I+
Sbjct 484 YEKF-----FHVKHAIEDIPIKTLNAWKKTKFNEREENEIKLPTPEKCSP--INLRIIQE 536
Query 255 LLGLTEPELISEQGGNAGTAVWWQG 279
+ L+ P+ + N +WW G
Sbjct 537 VEDLSTPQRLDSMLAN----IWWNG 557
> eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.55);
K01407 protease III [EC:3.4.24.55]
Length=962
Score = 85.9 bits (211), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 131/537 (24%), Positives = 226/537 (42%), Gaps = 41/537 (7%)
Query 22 LWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDR--NSIST 79
L V F + ++ + ++TYL+ G+L+ L+ GL +GIS D N S
Sbjct 290 LRVEFRIDNNSAKFRSKTDELITYLIGNRSPGTLSDWLQKQGLVEGISANSDPIVNGNSG 349
Query 80 LLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPS-SS 138
+L I LT KG A+R V+ IFSY+N LR+ G+ + +A IDF PS +
Sbjct 350 VLAISASLTDKGLANRDQVVAAIFSYLNLLREKGIDKQYFDELANVLDIDFRY--PSITR 407
Query 139 IMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKV 198
MD LA ++ H + ++ D + + L M+P A I + P
Sbjct 408 DMDYVEWLADTMIRVPVEHTLDAVNIADRYDAKAVKERLAMMTPQNARIWYISPKEPHNK 467
Query 199 DSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGLLGL 258
++ D Y V D A A+ A +P +IP + ++
Sbjct 468 TAYFVDAPYQV-----DKISAQTFADWQKKAADIALSLPELNPYIP--DDFSLIKSEKKY 520
Query 259 TEPELISEQGGNAGTAVWWQGQGAFAI-PRITVQLNGSILKD-KADLLSRTQGSLALAAI 316
PELI ++ + V + FA P+ V L IL++ KA +R Q + A+
Sbjct 521 DHPELIVDE---SNLRVVYAPSRYFASEPKADVSL---ILRNPKAMDSARNQ---VMFAL 571
Query 317 AEHLQEETVD-FQNCGVTHSLAFK---GTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMV 372
++L +D N ++F G + YT+ +L +L + + + +
Sbjct 572 NDYLAGLALDQLSNQASVGGISFSTNANNGLMVNANGYTQ-RLPQLFQALLEGYFSYTAT 630
Query 373 EPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFA 432
E ++ E+ K +++ AFE A+ A +L++ FSR + L + A
Sbjct 631 E-DQLEQAKSWYNQMMDSAEKGKAFEQAIMPAQMLSQVPYFSRDERRKILPSITLKEVLA 689
Query 433 KLNELKNVHVDAF-VMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVAS--LAMEQKQTIE 489
+ LK+ F V+GN+ Q+ T+ + +Q G D + + + +++KQ++
Sbjct 690 YRDALKSGARPEFMVIGNMTEAQATTLARDVQKQLG---ADGSEWCRNKDVVVDKKQSVI 746
Query 490 ATLANPIKGDKDHASL--VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGY 544
A G+ ++L V G ++L Q + Y+ LRTE QLGY
Sbjct 747 FEKA----GNSTDSALAAVFVPTGYDEYTSSAYSSLLGQIVQPWFYNQLRTEEQLGY 799
> dre:557565 nrd1, si:dkey-171o17.4; nardilysin (N-arginine dibasic
convertase) (EC:3.4.24.61)
Length=1061
Score = 85.5 bits (210), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 145/706 (20%), Positives = 285/706 (40%), Gaps = 90/706 (12%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGS----LAKRLRLLGLADGISPA-VDRN 75
+L +++ LP Y+ +P +++L+ + G GS L KR L L G S + D+N
Sbjct 343 ALTISWALPPQAKHYRVKPLHYISWLIGHEGVGSVLSLLRKRCWALSLFGGNSESGFDQN 402
Query 76 SISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQP 135
S ++ I + L+ +G + V+ IF Y+ L+ G + + + +FH +
Sbjct 403 STYSIFSISITLSDEGLQNFLQVIHIIFQYLKMLQSVGPQQRIYEEIQKIEANEFHYQEQ 462
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQL----LQEMSPSKAIIAFSD 191
+ I + A ++ N+ + H + GD L+ D +P ++ + L ++P KA I
Sbjct 463 TEPI-EFVANMSENMQLFPKEHFLCGDQLMFDFNPEASHCVISAALSLLTPGKANILLLS 521
Query 192 PDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKI 251
P E ++G Q+ V D+PQ A F + P L +P ++
Sbjct 522 PQHDGLCPLKEK--WFGTQYSVEDIPQEFRDLWA------GDFPLHPEL-QLPAENKFIA 572
Query 252 LPGLLGLTEP-------ELISEQGGNAGTAVWWQGQGAFAIPR--ITVQLNGSILKDKAD 302
L ++ ++I + G +W++ F IP+ QL +++
Sbjct 573 TDFTLRTSDCPDTDFPVKIIDNERGR----LWFRKDNKFKIPKAYARFQLLTPFIQESPK 628
Query 303 LLSRTQGSLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHM---GFEAYTEGQLSKLM 359
L L + +A +L E D + + ++L G + GF L ++
Sbjct 629 NL--VLFDLFVNIVAHNLAELAYDAEVAQLQYNLLPGDHGLFIRLKGFNHKLPLLLKLIV 686
Query 360 EHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLL 419
+H+A + P + E++K+ ++ P + + L+ IL + R ++
Sbjct 687 DHLADFSATPDVFN-MFIEQLKKTYYIILIRPE-RLGKDVRLQ---ILEHH----RWSVM 737
Query 420 NALQQTNYDDSFAKLNELKN-----VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHD 474
+ D S A L N + V+ V GN +S ++ F+E+ + P +
Sbjct 738 QKYEAIMADPSVADLMTFANRFKAELFVEGLVQGNFTSAESKEFLQCFIEKLKYAPHPIE 797
Query 475 DAVASLAMEQKQTIEATLANPI-KGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIY 533
V +E QT + K D + V +Q G+ ++ + + +L + +
Sbjct 798 PPVLFRVVELPQTHHLCKVQSLNKADANSEVTVYYQTGLKNLREHTLMELLVMHMEEPCF 857
Query 534 DSLRTEAQLGY-----------IAG---AKESQAASTALLQCFVEGA-----KAHPDEVV 574
D LRT+ LGY I G E+QA T FVEG + +++V
Sbjct 858 DFLRTKETLGYQVYPICRNTSGILGFSVTVETQA--TKFSTDFVEGKIEAFLVSFGEKLV 915
Query 575 KMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTK 634
++ DE L + E E + ++ ++ E+ F +
Sbjct 916 QLSDEAFGAQVTALIKLKECE---------------DTQLGDEVDRNWFEVVTQQYVFDR 960
Query 635 RDLEVKYLDNDFSRKQLLRTFAK-LSDPSRRMVVKLIADLEPEKEV 679
+ E++ L D ++ +L+ + + + SR++ + ++ E EKE+
Sbjct 961 LNKEIEIL-KDVTKDELVSFYMEHRKENSRKLSIHVVGFGEEEKEI 1005
> dre:565850 fk24c07; wu:fk24c07; K01411 nardilysin [EC:3.4.24.61]
Length=1091
Score = 76.3 bits (186), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 112/547 (20%), Positives = 219/547 (40%), Gaps = 48/547 (8%)
Query 21 SLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLR----LLGLADGIS-PAVDRN 75
+L + + LP Y+ +P + +L+ + G GS+ LR L L G S D+N
Sbjct 374 ALTITWALPPQEKHYRVKPLHYIAWLIGHEGTGSILSMLRRKCWALALFGGNSETGFDQN 433
Query 76 SISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQP 135
+ ++ I + LT +G + V +F Y+ L+ G + + + +FH +
Sbjct 434 TTYSIFSISITLTDEGFQNFYEVAHLVFQYLKMLQTLGPQQRIYEEIQKIEANEFHYQEQ 493
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
+ I + + N+ + + GD L+ + P + + L ++P KA + P+
Sbjct 494 TDPI-EYVEDICENMQLFPKEDFLTGDQLMFEFKPEVISAALNLLTPEKANLLLLSPEHE 552
Query 196 SKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRMPPSLMHIPKASELKILPGL 255
+ E ++G Q+ D+ QH A F + PSL H+P +E K +
Sbjct 553 GQCPLREK--WFGTQYSTEDIEQHWREIWA------KDFDLNPSL-HLP--AENKFIATD 601
Query 256 LGLT-----EPELISEQGGNAGTAVWWQGQGAFAIPRITVQ---LNGSILKDKADL-LSR 306
L + E N +W++ F IP+ V+ ++ + K +L L
Sbjct 602 FALKTSDCPDTEYPVRIMNNDRGCLWYKKDNKFKIPKAYVRFHLISPVVQKSPKNLVLFD 661
Query 307 TQGSLALAAIAEHLQEETV---DFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVA 363
++ + +AE E V +++ H L K GF+ +L L +
Sbjct 662 LFVNILVHNLAEPAYEADVAQLEYKLVAGEHGLVIKVKGFNH--------KLPLLFNLIV 713
Query 364 KLLSDPSMVEPERF----ERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLL 419
L+D S P+ F E++K+ ++ P L IL + + +
Sbjct 714 DYLADFSAA-PDVFSMFAEQLKKTYFNILIKPEKLGKDVRLL----ILEHSRWSTIQKYQ 768
Query 420 NALQQTNYDDSFAKLNELKN-VHVDAFVMGNIDRDQSLTMVEEFLEQAGFTPIDHDDAVA 478
L + D+ ++ K+ ++ + + GN+ +S+ ++ E+ F + + V
Sbjct 769 AVLDGLSVDELMEFVSGFKSELYAEGLLQGNVTSTESMGFLQYVTEKLQFKKLSVEVPVL 828
Query 479 SLAMEQKQTIEATLANPI-KGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLR 537
+E Q + KGD + V +Q G ++ + + +L + +D LR
Sbjct 829 FRVVELPQKHHLCKVKSLNKGDANSEVTVYYQSGPKNLREHTLMELLVMHMEEPCFDFLR 888
Query 538 TEAQLGY 544
T+ LGY
Sbjct 889 TKETLGY 895
> hsa:3416 IDE, FLJ35968, INSULYSIN; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=464
Score = 64.3 bits (155), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 87/436 (19%), Positives = 181/436 (41%), Gaps = 35/436 (8%)
Query 273 TAVWWQGQGAFAIPRITVQLNGSILKDKADLLSRTQGSLALAAIAEHLQEETVDFQNCGV 332
+ +W++ F +P+ + D L L L + + L E + G+
Sbjct 2 SKLWFKQDDKFFLPKACLNFEFFSPFAYVDPLHCNMAYLYLELLKDSLNEYAYAAELAGL 61
Query 333 THSLAFKGTGFHMGFEAYTEGQ---LSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVA 389
++ L G ++ + Y + Q L K++E +A ++ +RFE IK+ M+ +
Sbjct 62 SYDLQNTIYGMYLSVKGYNDKQPILLKKIIEKMATF-----EIDEKRFEIIKEAYMRSLN 116
Query 390 DPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAKLNEL-KNVHVDAFVMG 448
+ +HA+ +L A+++ +L AL A + +L +H++A + G
Sbjct 117 NFRAEQPHQHAMYYLRLLMTEVAWTKDELKEALDDVTLPRLKAFIPQLLSRLHIEALLHG 176
Query 449 NIDRDQSL---TMVEEFLEQAGFTPIDHDDAVASLAMEQKQTIEATLANPIKG------- 498
NI + +L MVE+ T I+H L + + E L P +G
Sbjct 177 NITKQAALGIMQMVED-------TLIEHAHTKPLLPSQLVRYREVQL--PDRGWFVYQQR 227
Query 499 DKDHASL---VQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAAS 555
++ H + + +Q + S + + L + Q ++ +++LRT+ QLGYI + +A
Sbjct 228 NEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLRTKEQLGYIVFSGPRRANG 287
Query 556 TALLQCFVEGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFS 615
L+ ++ K P + ++ L ++ + +M E + +A + S
Sbjct 288 IQGLRFIIQSEKP-PHYLESRVEAFLITMEKSIEDMTEEAFQKHIQALAIRRLDKPKKLS 346
Query 616 EDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKL--SDPSRRMVVKLIADL 673
+ K EI + F + + EV YL +++ +++ + ++ D RR V +
Sbjct 347 AECAKYWGEIISQQYNFDRDNTEVAYLKT-LTKEDIIKFYKEMLAVDAPRRHKVSVHVLA 405
Query 674 EPEKEVTLIGEAKAQD 689
++GE Q+
Sbjct 406 REMDSCPVVGEFPCQN 421
> cpv:cgd2_920 peptidase'insulinase-like peptidase'
Length=1028
Score = 62.0 bits (149), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 133/705 (18%), Positives = 273/705 (38%), Gaps = 103/705 (14%)
Query 16 FSSEPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRN 75
+ ++ L + F LP + + ++ + GEG ++ LR LA G S A+
Sbjct 276 YETDKRLKIYFPLPPLDKYNDSCAPAYIANIIGHKGEGGISSILRAKKLATGASFAITNE 335
Query 76 SISTLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQP 135
L V LT +G + G VL+ IF+++ + V ELV + F T QP
Sbjct 336 DPCALAQFGVVLTDEGYNNIGQVLEIIFNFLVLFKATPVIPELVDEFIGITRAGF-TYQP 394
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDA---DPRLTNQLLQEMSPSKAIIAFS-- 190
SI D + A L + + ++ D + +L+ +S I S
Sbjct 395 KFSIRDLFSLPAKYLKYKCKFEEILSSGFVVKKFSQDDVFS--ILEYLSNDNFFILLSSQ 452
Query 191 --DPDFTSKVDSFETDPYYGVQFRVLDLPQ-----------HHAVAMAVLTASPNAFRMP 237
+ ++ ++F + YYG ++ + +L + A+ + ++ PN F
Sbjct 453 AIEEEYKKNQENFIVEHYYGTKYSISELDEDLLSIINSSSPEKALKLGLILPKPNQFVST 512
Query 238 PSLMHIPK---ASELKILPGLLGLTEPELISEQGGNAGTAV-------WWQGQGAFAIPR 287
+ P+ ++ +P LL E + QG + W++ F P
Sbjct 513 DFSILNPQKVCVNDYLRIPELLNFDE---LKAQGNTDSYNIHSHPLNIWFKPDSTFNSPH 569
Query 288 ITVQLN---GSILKDKAD----LLSRTQGSLALAAIAEHLQE------------------ 322
+ + IL+ K + LS L E L E
Sbjct 570 SLINMRLVAERILEFKENSSFEKLSNFSNELVFQVFGEILNEVMYRSMHELSSDILAASL 629
Query 323 -ETVDF---------QNCGVTHSLAFKGTGFHMGFEAYTEG-QLSKLMEHVAKLLSDPSM 371
T++F Q G++H L + + + FE G ++ K + ++S
Sbjct 630 SYTINFNSRTNVFVLQGFGLSHKLNYLIS---IMFEKLYHGTEVRKYYDEAILIIS---- 682
Query 372 VEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSF 431
K + K++ TS++ E E+ + F+R++ LN L+ ++ F
Sbjct 683 ---------KDWKNKIIKPNLTSLSLECISESLSPF----FFNRQEKLNVLESFTFE-LF 728
Query 432 AKLNE--LKNVHVDAFVMGNIDRDQSLTM-VEEFLEQAGF-TPIDHDDAVASLAMEQ--- 484
+ + L N ++ +MGN + + ++ + F + ++ + +EQ
Sbjct 729 CSIRQHFLSNCRLEGLIMGNFSEPNAKCISIQHWKNLINFQNSVKNEVKSCGIKVEQFSI 788
Query 485 ---KQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQ 541
K+ I P DK+ ++ F LG ++ +V ++ F++ + LRT Q
Sbjct 789 VNLKKDIYTLNYIPNSSDKNGCWMLSFFLGEYNLRKQVLCDLILPFVSSEAFADLRTNQQ 848
Query 542 LGYIAGAKESQAASTALLQCFVEGAK-AHPDEVVKMIDEELSKAKEYLANMPEAEM-ARW 599
L Y+ A + ++ ++ +++ ++ + + ++++ ++K K L + EM +
Sbjct 849 LAYVVRATQIFSSPAIIIGYYLQSSEYTNALTLERLLEFHINKTKVELKSKLNKEMFIKL 908
Query 600 KEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDN 644
K++ L+ + +++K EI S F R ++ L+N
Sbjct 909 KDSTIQTLSSNPKSIFDEYKTYLHEINERSYLFDIRQRKIDILNN 953
> cpv:cgd2_930 peptidase'insulinase-like peptidase' ; K01408 insulysin
[EC:3.4.24.56]
Length=1013
Score = 62.0 bits (149), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 128/605 (21%), Positives = 237/605 (39%), Gaps = 112/605 (18%)
Query 19 EPSLWVAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSIS 78
E + F +P K P T +L + G GSL LR G +S ++ +
Sbjct 283 EKKVSFNFQIPDLRKFRKGLPEMYFTNILGHEGPGSLTSALRRNGWCLALSSGLNEMYSA 342
Query 79 TLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDF-HTTQPS- 136
L I + LT+KGA V++ +++N + + + E+VS + + S + F + +PS
Sbjct 343 NLFEIIITLTEKGAREVLSVIEYTLNFVNLVIKNEIDMEVVSDLEKLSQLVFDYRNRPSL 402
Query 137 -SSIMDEAARLAH-----NLLTYEPYHVVAGDSL-LIDADPRLTNQLLQEMSPSKAIIAF 189
+I + LA+ LLT+ G+ + +D D L Q P I
Sbjct 403 DETISNNVFALANLPPLKELLTF-------GNRVEKMDVDA--VKYLKQYFDPKNMFILL 453
Query 190 SDPDFTSKV------DSFETDPYYGVQFRVLDL-PQHHAVA--MAVLTASPNAFRMPPSL 240
S P+ + + D D +Y + + L+ P+ + +++ AS +MP
Sbjct 454 SIPENKALIEDERLKDKLIYDRHYNINYLKLEFGPEIKEIISNISLSNASRFGLKMPTKN 513
Query 241 MHIPKASELKILPGLLGLTEPELISEQGGNAGTAV--WWQGQGAFAIP------------ 286
+IP+ +L G P ++ G + V +++ F P
Sbjct 514 NYIPENFDLMNTYGGNMQAFPTILEIPGNSFSDRVVAYYKPDTNFQTPHGFSQFFFFSSS 573
Query 287 RITVQLNGSILKDKADLLSRTQGSLALAA-IAEHLQEETVDFQNCGVTHSLAFKGTGFHM 345
++T +L L+ T SL L+ +AE T+ + ++ + + M
Sbjct 574 KVTCEL----------LVLDTLTSLTLSKFVAEEAYNATIANLDYKISGGYNLRNS---M 620
Query 346 GFEAYTEGQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAA 405
+ T + M + K L S+VE + +KQ+ FE ALE +
Sbjct 621 NCLSITISGFNDKMHTLLKFLI-KSLVE---LKNDGKKQL-------YKSFFEDALEESR 669
Query 406 ILTRNDAF----------------------SRKDLLNALQQTNYD---DSFAKLNELKNV 440
+ RN F S++++L+ L T Y+ D +
Sbjct 670 LSVRNSLFNPDILAHLTSYNFREFYSVYTPSKEEILSILSTTTYERLCDHISTF--FSQC 727
Query 441 HVDAFVMGNIDRDQSLTMVE-----EFLEQAGFTP----------IDHDDAVAS--LAME 483
+ + +GN++++Q+ +VE E L ID + A+ S
Sbjct 728 LIKSITVGNLNKEQARELVETVTIKELLSSEQLNSKMEKTIIRNCIDLEKAIESDPEIKS 787
Query 484 QKQTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLG 543
+ + ++ NP+ DK+ + + +G ++ + V L +L+++L+ Y LRT QLG
Sbjct 788 NRIILSKSVINPM--DKNGSVIYSIDMGEYNLRNYVLLELLSKYLDSNCYLELRTNQQLG 845
Query 544 YIAGA 548
YI A
Sbjct 846 YIVHA 850
> tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56)
Length=941
Score = 53.1 bits (126), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 47/182 (25%), Positives = 79/182 (43%), Gaps = 8/182 (4%)
Query 26 FGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGIKV 85
F LP +++ +P ++ +LE+ G SL+ +L+ GL + + T+L + V
Sbjct 292 FYLPPQAKNWRSKPLQFISEMLEHEGPTSLSSKLKREGLITSLVTDYWSPELCTVLQVNV 351
Query 86 DLTQKGAAHRGLVLQEIFSYINFLRDHGVGHE---LVSTMAQQSHIDF-HTTQPSSSIMD 141
LT+ G + V + + FLR+ GV V+ MA+ + F P +
Sbjct 352 RLTEGGRSKES-VYKIGHALFTFLRNLGVSRPERWRVTEMAKIRQLGFAFADMPDPYAL- 409
Query 142 EAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQE-MSPSKAIIAFSDPDFTSKVDS 200
R L Y P V+AGD L+ DP + Q +Q+ + P + D + VD
Sbjct 410 -TVRAVEGLNYYTPEEVIAGDRLIYHFDPDIIQQYVQKFLVPDNVRLFIFDKKLAADVDR 468
Query 201 FE 202
E
Sbjct 469 EE 470
> tgo:TGME49_069870 hypothetical protein
Length=413
Score = 52.0 bits (123), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 77/149 (51%), Gaps = 19/149 (12%)
Query 484 QKQTIEATLANPIKGDKDHASLVQFQLG-IPSIEDRVNLAVLTQFLNRRIYDSLRTEAQL 542
Q ++I L NP DK + + + ++G +P+I DR L ++++++++R ++ LRTE QL
Sbjct 190 QLRSIRKNL-NP--NDKKNQAYLLIEVGALPNIHDRAVLYMVSRWMSQRFFNKLRTEQQL 246
Query 543 GYIAGAKESQAASTALLQCFVEGAKAHP----DEVVKMIDEELSK--AKEYLANMPEAEM 596
GY+ S+ + F+ + P D +V+ I+ E SK +E A + +A +
Sbjct 247 GYLTAMHSSRLEDRFYYRFFIT-STYDPAEVADRIVEFINAERSKIPTQEEFATLKQAAI 305
Query 597 ARWKEAAHAKLTKMEANFSEDFKKSAEEI 625
WK+ N E+F+K+ ++
Sbjct 306 DVWKQKPK--------NIFEEFRKNRRQV 326
> cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=856
Score = 52.0 bits (123), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/130 (27%), Positives = 65/130 (50%), Gaps = 6/130 (4%)
Query 24 VAFGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGI 83
+ F P + QP + +L+ + G GSL L+ LG IS D ++I++ G+
Sbjct 279 IKFPFPDLNGEFLSQPGDYIAHLIGHEGPGSLLSELKRLGWV--ISLEADNHTIASGFGV 336
Query 84 ---KVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIM 140
+DL+ +G H V+Q +F++I FL+ G + +A+ + +DF + M
Sbjct 337 FSVTMDLSTEGLEHVDDVIQLVFNFIGFLKSSGPQKWIHDELAELNAVDFRFDDVKHT-M 395
Query 141 DEAARLAHNL 150
++A+ LA L
Sbjct 396 EKASILAECL 405
Score = 48.1 bits (113), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 93/446 (20%), Positives = 173/446 (38%), Gaps = 40/446 (8%)
Query 227 LTASPNAFRMPPSLMHIPKASELKIL-PGLLGLTEPELISEQGGNAGTAVWWQGQGAFAI 285
L S +AF +P +IP + K P G P LISE + VW++ +
Sbjct 417 LKTSHHAFNLPEKNEYIPSKFDQKPREPVKSGY--PRLISE---DEWIQVWFKQDNEYNS 471
Query 286 PRITVQLNGSILKDKADLLSRTQGSLALAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHM 345
P+ G + L+++ ++ + + EET + + G+ +G +
Sbjct 472 PK-----QGIMFALTTPLVAKKSKNVVAFKSLDTIIEETYNARLAGLECQFESSSSGVQI 526
Query 346 GFEAYTEGQLSKLMEHVAKLLSD---PSMVEPERFERIKQKQMKLVADPATSMAFE--HA 400
Y E Q S +H+ +++ + FE +K+ T+ AF H
Sbjct 527 RVFGYDEKQ-SLFAKHLVNRMANFQVNRLCFDISFESLKR--------TLTNHAFSQPHD 577
Query 401 LEAAAI--LTRNDAFSRKDLLNALQQTNYDD--SFAKLNELKNVHVDAFVMGNIDRDQSL 456
L A I L ++ +S++ LL +D FA + L+ H++ FV GN +L
Sbjct 578 LSAHFIDLLVVDNIWSKEQLLAVCDSVTLEDVHGFA-IKMLQAFHMELFVHGNSTEKDTL 636
Query 457 TMVEEFLE-----QAGFTPIDHDDAVASLAMEQKQTIEATLANPIKGDKDHASLVQFQLG 511
+ +E + P+ D+ ++ E + K V FQ+G
Sbjct 637 QLSKELSDILKSVAPNSRPLKRDEHNPHRELQLINGHEHVYRHFQKTHDVGCVEVAFQIG 696
Query 512 IPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGAKAHPD 571
+ S + +L + + Y LRT LGY + L V+G ++ D
Sbjct 697 VQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLNDGNVYLHVIVQGPES-AD 755
Query 572 EVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNC 631
V++ I+ L A+E + MP+ + A + S+ F EI +
Sbjct 756 HVLERIEVFLESAREEIVAMPQEDF---DYQVWAMFKENPPTLSQCFSMFWSEIHSRQYN 812
Query 632 FTKRDLEVKYLDNDFSRKQLLRTFAK 657
F R+ EV+ + ++++++ F +
Sbjct 813 FG-RNKEVRGISKRITKEEVINFFDR 837
> tgo:TGME49_044490 insulin-degrading enzyme, putative (EC:3.4.24.56)
Length=592
Score = 50.4 bits (119), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 58/274 (21%), Positives = 107/274 (39%), Gaps = 44/274 (16%)
Query 412 AFSRKDLLNALQQTNYDDSFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFLEQ------ 465
S +DLL L+QT D + V+ V+GNI + MVE L+
Sbjct 193 CLSYEDLLRVLEQTTLDVQEVPKTLFERACVEGLVVGNISSAEVCVMVEMALKNLNIETT 252
Query 466 --------------------------AGFTPIDHDD---AVASLAMEQKQTIEATLA--- 493
+G +D + +L E+ + E L
Sbjct 253 LDSNSVPEKAVVDLASLDLARLRSSGSGVAGLDEEREAMQCRTLVCEELKASEVKLKTRS 312
Query 494 -NPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYIAGAKESQ 552
N D++ + ++FQLG +R L++ + +++ +D LRT+ QL Y+ A S
Sbjct 313 ENSNTQDRNSVAFLRFQLGNLEDRERSMLSLFSHCISQAFFDDLRTQQQLDYVLHAHRSF 372
Query 553 AASTALLQCFVEGAKAHPD----EVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLT 608
+ + FV G+ D + +++++ LS + A +P+A + + A ++L
Sbjct 373 QLRSQGMHFFVAGS-TFSDLMTLRIGRLVEKYLSSEQGLHAGLPDALCEKHRSALVSELR 431
Query 609 KMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYL 642
N E+ ++ EI F + D + L
Sbjct 432 VRPQNAFEEAQRYTREISTWYFMFNRHDRTIAEL 465
> cpv:cgd3_4270 peptidase'insulinase like peptidase' ; K01408
insulysin [EC:3.4.24.56]
Length=1176
Score = 45.1 bits (105), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 37/151 (24%), Positives = 64/151 (42%), Gaps = 4/151 (2%)
Query 46 LLEYTGEGSLAKRLRLLGLA-DGISPAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFS 104
L+ + GSL L+ L D +D + T I +LT G + G +L FS
Sbjct 354 LISFDRPGSLGHHLKSKKLILDVYFSLIDDDLGFTNAVIGFELTIDGEKNIGYILLSFFS 413
Query 105 YINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYE--PYHVVAGD 162
I F ++ E+ + +I F P+S+ D+ + + YE P V+ D
Sbjct 414 AIKFASNNEFSKEIYDEWRKLLYISFKYEDPTST-FDQCKEIVTYYIQYECKPEDVLYSD 472
Query 163 SLLIDADPRLTNQLLQEMSPSKAIIAFSDPD 193
+ + DP + ++ +++P II PD
Sbjct 473 YYMDEFDPNIYKEINSQLTPENLIITLERPD 503
> cpv:cgd3_4260 peptidase'insulinase like peptidase' ; K01408
insulysin [EC:3.4.24.56]
Length=1172
Score = 41.2 bits (95), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 36/151 (23%), Positives = 63/151 (41%), Gaps = 4/151 (2%)
Query 46 LLEYTGEGSLAKRLRLLGLADGIS-PAVDRNSISTLLGIKVDLTQKGAAHRGLVLQEIFS 104
L+ + GSL L+ L + +D N T I +LT G + G +L FS
Sbjct 350 LISFDRPGSLGHHLKSKKLILNMDFSIIDDNLGFTNAVIGFELTIDGEKNIGYILLSFFS 409
Query 105 YINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAARLAHNLLTYE--PYHVVAGD 162
I F ++ E+ I F P+S+ D++ + + +E P V+ D
Sbjct 410 VIKFASNNEFSKEIYDEWKNLIDISFKYEDPTST-SDQSEEIVTYYIKHECKPEDVLYSD 468
Query 163 SLLIDADPRLTNQLLQEMSPSKAIIAFSDPD 193
+ + DP + ++ +++P II PD
Sbjct 469 YYMDEFDPNIYKEINSQLTPENLIITLERPD 499
> cpv:cgd3_4170 secreted insulinase like peptidase
Length=1289
Score = 40.4 bits (93), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 33/169 (19%), Positives = 71/169 (42%), Gaps = 3/169 (1%)
Query 26 FGLPATLTSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGIKV 85
F + + ++K+ PT + YLL+ +G L K L+ +G+++ I V + L I +
Sbjct 328 FPIEIQVVNWKRIPTMYIKYLLDGNYKGILRKYLKSIGISNPIKVGVVNYEGFSTLDISI 387
Query 86 DLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSSSIMDEAAR 145
DL H +++ + S + ++ + V +V + I F+ + + D A
Sbjct 388 DLYNSQLRHSWNLVKAVISAVKYIIELPVSERIVEEAKNVADIIFNYRETEFT-RDLAYN 446
Query 146 LAHNLLTY--EPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDP 192
+ + Y +P ++ D ++ D + + + I F P
Sbjct 447 IVYKASKYRIKPQEIIYADEVMEIVDISFIKAFISSIKIDQVSIFFFTP 495
Score = 34.7 bits (78), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 44/159 (27%), Positives = 69/159 (43%), Gaps = 12/159 (7%)
Query 320 LQEETVDFQNCGVTHSLA----FKGTGFHMGFEAYTEG---QLSKLMEHVAKLLSDPS-M 371
L++E DF+ ++ + G GFE + +G L K + AK LS P+
Sbjct 829 LEDEIYDFRIARNEFKISSFNDYTYNGLPNGFEIHLKGFHDVLPKFLRIFAKHLSHPNKY 888
Query 372 VEPERFE-RIKQKQMKLVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDS 430
E+FE K L + + +L +T N + S D LN L+ +Y D
Sbjct 889 FTLEQFEDAFKHVNRYLYQYVYFTPSIIKSLTILRSVTENQSLSPFDRLNELKYISYRD- 947
Query 431 FAKLNEL--KNVHVDAFVMGNIDRDQSLTMVEEFLEQAG 467
+L+E K V+ M NID ++ +V EFL+ G
Sbjct 948 IIELSEFFAKQGQVEGLFMNNIDPFEAGLVVNEFLDCLG 986
> tgo:TGME49_114850 hypothetical protein
Length=2136
Score = 36.6 bits (83), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 57/254 (22%), Positives = 92/254 (36%), Gaps = 59/254 (23%)
Query 46 LLEYTGEGSLAKRLRLLGLADGISPAVDRNSISTLLGIKVDLTQKGAAHRGLVL--QEIF 103
+L G GSL LR GLA+ I + + + L V+L H ++L +F
Sbjct 869 VLGSEGPGSLVAHLRKQGLAESIDVEAEESRCYSTLRASVELKDFNMNHTAIMLIGSSLF 928
Query 104 SYINFLRDHGVGHELVSTMAQQSHIDFHT----------------TQPSS---------- 137
SY+ F+R+ + V + + + F T P+
Sbjct 929 SYLRFIRESVLDRAFVESRMRMYRLAFDAHVPPLPPRPPLPGALGTIPALLPSSLLQSSL 988
Query 138 ------------SIMDEAARLAHNLL-TYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSK 184
S +E A LA +L P V LL D + L +++P
Sbjct 989 PSSSPGQAGTNFSPQEEVAYLAADLQEILSPARVFTEQWLLEDVGKQSLEHYLDQLTPDN 1048
Query 185 AIIAFSDP---DFTSKVDSFETDPYYGVQFRV--LDLPQHHAVAM--------AVLTASP 231
I+ S P + V SF +G+ F + LD Q A + A+L A
Sbjct 1049 LILMISSPLVVPLCTLVSSF-----FGIPFAINPLDATQDAAWSALVSLPPERALLLARR 1103
Query 232 NAFRMPPSLMHIPK 245
F +PP+ + +P+
Sbjct 1104 TGFSLPPASLFVPE 1117
> cpv:cgd6_5520 peptidase'insulinase like peptidase' ; K01408
insulysin [EC:3.4.24.56]
Length=570
Score = 34.3 bits (77), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/129 (19%), Positives = 58/129 (44%), Gaps = 8/129 (6%)
Query 79 TLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQ---QSHIDFHTTQP 135
L I ++LT G + +L+EI+S I +++++ +++ + + ++ P
Sbjct 369 NLFKIYIELTINGIKNIEYILKEIYSAIIYIKENISFEQILQDYNHSQNEQYYNYVDDSP 428
Query 136 SSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFT 195
++ I+D+ + L+ P +V+ + + N +L E+ P +I + F
Sbjct 429 NNQIIDKY--FNYKLM---PKYVIINNIETNQINENTINSILSEIEPENMLILINTNKFN 483
Query 196 SKVDSFETD 204
D FE +
Sbjct 484 KLFDHFENN 492
> dre:100073330 pts, zgc:162276; 6-pyruvoyltetrahydropterin synthase
(EC:4.2.3.12); K01737 6-pyruvoyl tetrahydrobiopterin
synthase [EC:4.2.3.12]
Length=139
Score = 33.9 bits (76), Expect = 2.6, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 40/84 (47%), Gaps = 10/84 (11%)
Query 402 EAAAILTRNDAFSRKDLLNA--LQQTNYDDSFAKLNELK----NVHVDAFVMGNIDRDQS 455
E A +TR +FS L++ L +F K N N V+ V G ID++
Sbjct 3 ERVAFITRVCSFSACHRLHSKCLSDEENKRTFGKCNNPNGHGHNYTVEVTVRGKIDKNTG 62
Query 456 LTM----VEEFLEQAGFTPIDHDD 475
+ M ++EF+E+A P+DH +
Sbjct 63 MVMNLTDLKEFIEEAVMKPLDHKN 86
> ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein
Length=820
Score = 33.5 bits (75), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 48/223 (21%), Positives = 86/223 (38%), Gaps = 28/223 (12%)
Query 428 DDSFAKLNELKNVHVDAFVMGNIDRDQSLTMVEEFL--EQAGFTPIDHDDAVASLAMEQK 485
+D F++ E +V ++G + FL +++G P DA+ +A+
Sbjct 576 EDMFSQTKERNSVTYTTMILGYGQHGMGERAISLFLSMQESGIKP----DAITFVAVLSA 631
Query 486 QTIEATLANPIKGDKDHASLVQFQLGIPSIEDRVNLAVLTQFLNRRIYDSLRTEAQLGYI 545
+ + +K ++ + Q PS E + +T L R R ++
Sbjct 632 CSYSGLIDEGLKIFEEMREVYNIQ---PSSE---HYCCITDMLGR----VGRVNEAYEFV 681
Query 546 AGAKESQAASTALLQCFVEGAKAHPD-EVVKMIDEELSK---AKEY------LANMPEAE 595
G E + L + K H + E+ + + E L+K K + L+NM AE
Sbjct 682 KGLGE-EGNIAELWGSLLGSCKLHGELELAETVSERLAKFDKGKNFSGYEVLLSNM-YAE 739
Query 596 MARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLE 638
+WK + E ++ +S EI + NCF RD E
Sbjct 740 EQKWKSVDKVRRGMREKGLKKEVGRSGIEIAGYVNCFVSRDQE 782
> ath:AT5G18240 MYR1; MYR1 (MYb-related protein 1); transcription
factor
Length=402
Score = 32.7 bits (73), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 40/83 (48%), Gaps = 14/83 (16%)
Query 313 LAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSMV 372
L +I E QE T+ QN G G EA T+ QLS+L+ V+ D S +
Sbjct 182 LQSILEKAQE-TLGRQNLGAA------------GIEA-TKAQLSELVSKVSADYPDSSFL 227
Query 373 EPERFERIKQKQMKLVADPATSM 395
EP+ + + +QM+ P +S+
Sbjct 228 EPKELQNLHHQQMQKTYPPNSSL 250
> cpv:cgd3_4190 secreted insulinase like peptidase
Length=1085
Score = 32.7 bits (73), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 40/66 (60%), Gaps = 3/66 (4%)
Query 78 STLLGIKVDLTQKGAAHRGLVLQEIFSYINFLRDHGVGHELVSTMAQQSHIDFHTTQPSS 137
+T+L ++V+LT+KG + ++++ I SY+N ++ L + Q+ ++H T+ S+
Sbjct 377 TTILYLEVNLTKKGLQNIPIIIESIASYLNLIKRTVASDRLFA--EAQNLFNYHLTK-ST 433
Query 138 SIMDEA 143
I+ EA
Sbjct 434 VILSEA 439
> xla:444677 MGC84289 protein; K01587 phosphoribosylaminoimidazole
carboxylase / phosphoribosylaminoimidazole-succinocarboxamide
synthase [EC:4.1.1.21 6.3.2.6]
Length=425
Score = 32.0 bits (71), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 2/53 (3%)
Query 26 FGLPATL--TSYKKQPTSVLTYLLEYTGEGSLAKRLRLLGLADGISPAVDRNS 76
+GLP L TS K P L EY G+G + + G ++G+ P + NS
Sbjct 291 YGLPCELRVTSAHKGPDETLRIKSEYEGDGMATVFIAVAGRSNGLGPVMSGNS 343
Lambda K H
0.317 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 39206744972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40