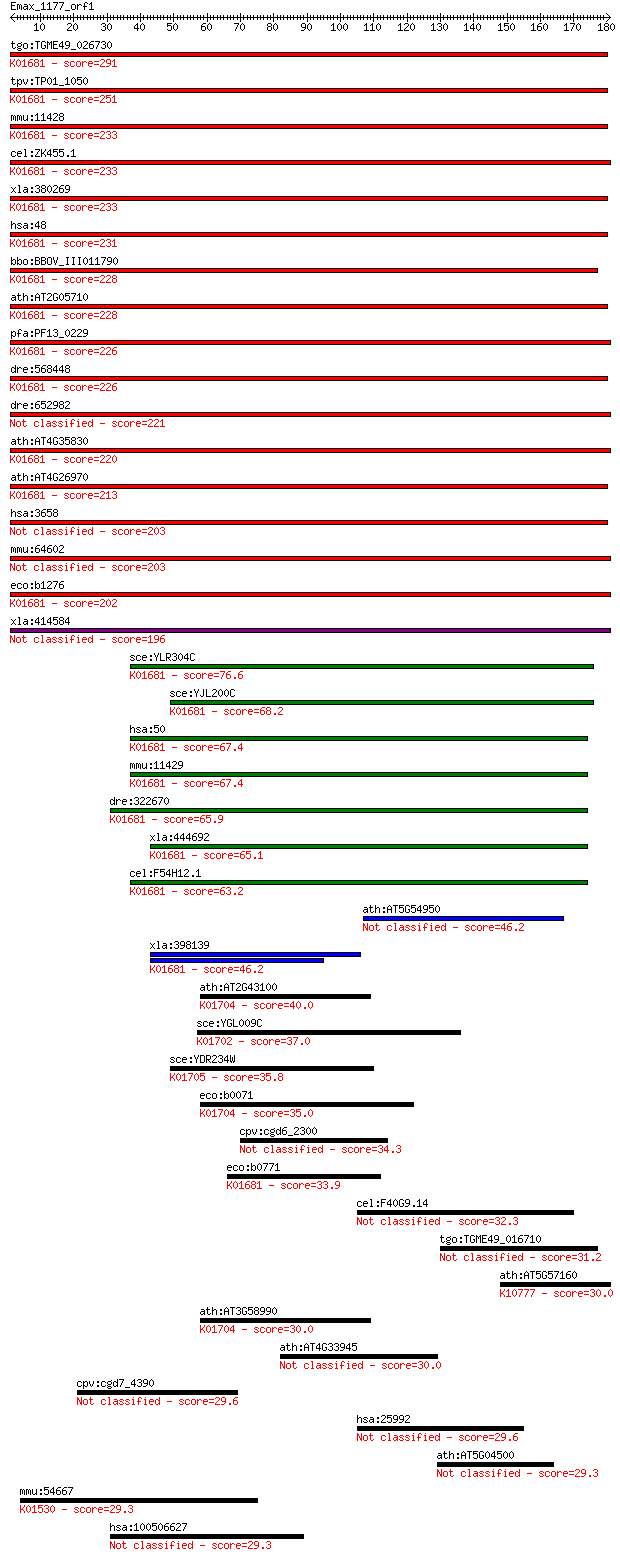
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1177_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_026730 aconitate hydratase, putative (EC:4.2.1.3); ... 291 6e-79
tpv:TP01_1050 aconitate hydratase; K01681 aconitate hydratase ... 251 7e-67
mmu:11428 Aco1, AI256519, Aco-1, Irebp, Irp1; aconitase 1 (EC:... 233 2e-61
cel:ZK455.1 aco-1; ACOnitase family member (aco-1); K01681 aco... 233 3e-61
xla:380269 aco1, MGC53330, ratireb; aconitase 1, soluble (EC:4... 233 3e-61
hsa:48 ACO1, ACONS, IREB1, IREBP, IREBP1, IRP1; aconitase 1, s... 231 1e-60
bbo:BBOV_III011790 17.m08005; aconitate hydratase 1 family pro... 228 9e-60
ath:AT2G05710 aconitate hydratase, cytoplasmic, putative / cit... 228 9e-60
pfa:PF13_0229 aconitase (EC:4.2.1.3); K01681 aconitate hydrata... 226 3e-59
dre:568448 aco1, wu:fo75b03; aconitase 1, soluble (EC:4.2.1.3)... 226 4e-59
dre:652982 ireb2, im:7153062; iron-responsive element binding ... 221 8e-58
ath:AT4G35830 aconitate hydratase, cytoplasmic / citrate hydro... 220 2e-57
ath:AT4G26970 aconitate hydratase/ copper ion binding (EC:4.2.... 213 2e-55
hsa:3658 IREB2, ACO3, FLJ23381, IRP2, IRP2AD; iron-responsive ... 203 3e-52
mmu:64602 Ireb2, D9Ertd85e, DKFZp564D1164, Irp2; iron responsi... 203 3e-52
eco:b1276 acnA, acn, ECK1271, JW1268; aconitate hydratase 1 (E... 202 4e-52
xla:414584 ireb2, MGC83131; iron-responsive element binding pr... 196 5e-50
sce:YLR304C ACO1, GLU1; Aco1p (EC:4.2.1.3); K01681 aconitate h... 76.6 4e-14
sce:YJL200C ACO2; Aco2p (EC:4.2.1.3); K01681 aconitate hydrata... 68.2 1e-11
hsa:50 ACO2, ACONM, MGC20605, MGC33908; aconitase 2, mitochond... 67.4 2e-11
mmu:11429 Aco2, Aco-2, Aco3, D10Wsu183e; aconitase 2, mitochon... 67.4 3e-11
dre:322670 aco2, cb1017, wu:fa10e03, wu:fb69g04, wu:fc20c11; a... 65.9 7e-11
xla:444692 aco2, MGC84375; aconitase 2, mitochondrial (EC:4.2.... 65.1 1e-10
cel:F54H12.1 aco-2; ACOnitase family member (aco-2); K01681 ac... 63.2 5e-10
ath:AT5G54950 aconitate hydratase-related / citrate hydro-lyas... 46.2 5e-05
xla:398139 aconitase; K01681 aconitate hydratase 1 [EC:4.2.1.3] 46.2 5e-05
ath:AT2G43100 aconitase C-terminal domain-containing protein (... 40.0 0.004
sce:YGL009C LEU1; Isopropylmalate isomerase, catalyzes the sec... 37.0 0.034
sce:YDR234W LYS4, LYS3; Homoaconitase, catalyzes the conversio... 35.8 0.091
eco:b0071 leuD, ECK0073, JW0070; 3-isopropylmalate dehydratase... 35.0 0.14
cpv:cgd6_2300 Cut4/Apc1p/TSG24 family protein; meiotic check p... 34.3 0.23
eco:b0771 ybhJ, ECK0760, JW5103; predicted hydratase; K01681 a... 33.9 0.29
cel:F40G9.14 hypothetical protein 32.3 0.91
tgo:TGME49_016710 hypothetical protein 31.2 2.2
ath:AT5G57160 ATLIG4; ATLIG4; DNA ligase (ATP)/ protein bindin... 30.0 4.5
ath:AT3G58990 aconitase C-terminal domain-containing protein (... 30.0 4.7
ath:AT4G33945 armadillo/beta-catenin repeat family protein 30.0 5.0
cpv:cgd7_4390 NudC ortholog 29.6 5.5
hsa:25992 SNED1, DKFZp586B2420, FLJ00133, SST3, Snep; sushi, n... 29.6 6.0
ath:AT5G04500 glycosyltransferase family protein 47 29.3
mmu:54667 Atp8b2, Id; ATPase, class I, type 8B, member 2 (EC:3... 29.3 7.0
hsa:100506627 DCDC5, DKFZp779D068, FLJ44078, FLJ46154, KIAA149... 29.3 7.8
> tgo:TGME49_026730 aconitate hydratase, putative (EC:4.2.1.3);
K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=1055
Score = 291 bits (746), Expect = 6e-79, Method: Compositional matrix adjust.
Identities = 130/179 (72%), Positives = 162/179 (90%), Gaps = 0/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGNDE+MVRGTFANIRLVNK+CP GPK++H+P+ E+LPV +VA +Y+AE +PMIVL
Sbjct 869 GARRGNDEIMVRGTFANIRLVNKLCPKDGPKSVHVPSGEVLPVYDVAMKYKAERKPMIVL 928
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYGSGSSRDWAAKGPYL GVKA+IAESFERIHR+NLVGMGILPLQF EGQ+A++L L
Sbjct 929 AGKEYGSGSSRDWAAKGPYLMGVKAIIAESFERIHRTNLVGMGILPLQFQEGQNAESLGL 988
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
TG+E+FNI+LN G ++PGS++ V T GK+F+ +CRIDT++EV+YF+NGG LHYVLRN+
Sbjct 989 TGKEQFNISLNKGEIIPGSLMTVKTSDGKAFDVRCRIDTELEVKYFQNGGILHYVLRNL 1047
> tpv:TP01_1050 aconitate hydratase; K01681 aconitate hydratase
1 [EC:4.2.1.3]
Length=912
Score = 251 bits (642), Expect = 7e-67, Method: Compositional matrix adjust.
Identities = 113/179 (63%), Positives = 146/179 (81%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND+VM RGTFANIR+ N +CP GP +H PTN+++ V + +E Y+ + P++V+
Sbjct 731 GSRRGNDKVMSRGTFANIRINNLLCPGQGPNTVHFPTNKLMSVYDASELYQRDNTPLVVV 790
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYG+GSSRDWAAKGP L GVKA++AESFERIHR+NLVG GILPLQFL+GQ+A TL L
Sbjct 791 AGKEYGTGSSRDWAAKGPLLLGVKAILAESFERIHRTNLVGCGILPLQFLDGQNATTLNL 850
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
TG E+F + L G +VPGS+VRV+TD G SF+ KCR+DT +E EY+++GG L YVLR++
Sbjct 851 TGTEKFTVHL-GSDVVPGSLVRVTTDTGLSFDTKCRVDTQIESEYYKHGGILQYVLRSI 908
> mmu:11428 Aco1, AI256519, Aco-1, Irebp, Irp1; aconitase 1 (EC:4.2.1.3);
K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=889
Score = 233 bits (595), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 110/179 (61%), Positives = 139/179 (77%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND +M RGTFANIRL+NK P+ +HLP+ E L V + AERY+ G P+IVL
Sbjct 710 GSRRGNDAIMARGTFANIRLLNKFLNKQAPQTVHLPSGETLDVFDAAERYQQAGLPLIVL 769
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYGSGSSRDWAAKGP+L G+KAV+AES+ERIHRSNLVGMG++PL++L G++AD+L L
Sbjct 770 AGKEYGSGSSRDWAAKGPFLLGIKAVLAESYERIHRSNLVGMGVIPLEYLPGETADSLGL 829
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
TGRER+ I + L P V++ D GK+F+A R DTDVE+ YF NGG L+Y++R M
Sbjct 830 TGRERYTINIPED-LKPRMTVQIKLDTGKTFQAVMRFDTDVELTYFHNGGILNYMIRKM 887
> cel:ZK455.1 aco-1; ACOnitase family member (aco-1); K01681 aconitate
hydratase 1 [EC:4.2.1.3]
Length=887
Score = 233 bits (593), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 106/180 (58%), Positives = 139/180 (77%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGNDE+M RGTFANIRLVNK+ GP +H+P+ E L + + A++Y+ G P I+L
Sbjct 708 GARRGNDEIMARGTFANIRLVNKLASKVGPITLHVPSGEELDIFDAAQKYKDAGIPAIIL 767
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYG GSSRDWAAKGP+LQGVKAVIAESFERIHRSNL+GMGI+P Q+ GQ+AD+L L
Sbjct 768 AGKEYGCGSSRDWAAKGPFLQGVKAVIAESFERIHRSNLIGMGIIPFQYQAGQNADSLGL 827
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
TG+E+F+I + L PG ++ V+ G F+ CR DT+VE+ Y+RNGG L Y++R ++
Sbjct 828 TGKEQFSIGV-PDDLKPGQLIDVNVSNGSVFQVICRFDTEVELTYYRNGGILQYMIRKLI 886
> xla:380269 aco1, MGC53330, ratireb; aconitase 1, soluble (EC:4.2.1.3);
K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=891
Score = 233 bits (593), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 111/179 (62%), Positives = 137/179 (76%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND VM RGTFANIRL NK P ++ P+NE L + + AERY+ EG +I+L
Sbjct 710 GSRRGNDAVMARGTFANIRLFNKFINKQSPLTIYFPSNETLDIFDAAERYQNEGHNLILL 769
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
GKEYGSGSSRDWAAKGP+LQG+KAV+AES+ERIHRSNLVGMGI+PLQ+L G+SA+ L L
Sbjct 770 TGKEYGSGSSRDWAAKGPFLQGIKAVLAESYERIHRSNLVGMGIIPLQYLPGESAEALGL 829
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
+GRER+ I + L PG V + D GKSF+A R DTDVE+ Y+RNGG L+Y++R M
Sbjct 830 SGRERYTIIIPED-LRPGMNVEIKLDTGKSFDAIMRFDTDVELTYYRNGGILNYMIRKM 887
> hsa:48 ACO1, ACONS, IREB1, IREBP, IREBP1, IRP1; aconitase 1,
soluble (EC:4.2.1.3); K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=889
Score = 231 bits (588), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 110/179 (61%), Positives = 139/179 (77%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND VM RGTFANIRL+N+ P+ +HLP+ EIL V + AERY+ G P+IVL
Sbjct 710 GSRRGNDAVMARGTFANIRLLNRFLNKQAPQTIHLPSGEILDVFDAAERYQQAGLPLIVL 769
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYG+GSSRDWAAKGP+L G+KAV+AES+ERIHRSNLVGMG++PL++L G++AD L L
Sbjct 770 AGKEYGAGSSRDWAAKGPFLLGIKAVLAESYERIHRSNLVGMGVIPLEYLPGENADALGL 829
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
TG+ER+ I + L P V+V D GK+F+A R DTDVE+ YF NGG L+Y++R M
Sbjct 830 TGQERYTIII-PENLKPQMKVQVKLDTGKTFQAVMRFDTDVELTYFLNGGILNYMIRKM 887
> bbo:BBOV_III011790 17.m08005; aconitate hydratase 1 family protein
(EC:4.2.1.3); K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=908
Score = 228 bits (581), Expect = 9e-60, Method: Compositional matrix adjust.
Identities = 105/177 (59%), Positives = 139/177 (78%), Gaps = 3/177 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGNDE+MVRGTFANIRL N +CP+ GPK + PT E+L + + +E+Y+ +G P++V+
Sbjct 729 GSRRGNDEIMVRGTFANIRLSNLLCPNQGPKTVFHPTGEVLSIFDASEKYKQQGTPLVVV 788
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYGSGSSRDWAAKGP L G++A+ AESFERIHR+NLVG GILPLQF+ G++A ++ +
Sbjct 789 AGKEYGSGSSRDWAAKGPALLGIRAIFAESFERIHRTNLVGFGILPLQFMPGENAASVGI 848
Query 121 TGRERFNIALNG-GRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVL 176
TGRE+F I +G +L PG V V D G F +CRIDT +E++Y+++GG L YVL
Sbjct 849 TGREKFTI--DGLDKLSPGCQVEVVADTGIKFNMRCRIDTALELQYYQHGGILQYVL 903
> ath:AT2G05710 aconitate hydratase, cytoplasmic, putative / citrate
hydro-lyase/aconitase, putative (EC:4.2.1.3); K01681
aconitate hydratase 1 [EC:4.2.1.3]
Length=990
Score = 228 bits (581), Expect = 9e-60, Method: Compositional matrix adjust.
Identities = 115/182 (63%), Positives = 137/182 (75%), Gaps = 3/182 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNK-MCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGNDE+M RGTFANIR+VNK M GPK +H+P+ E L V + A RY++ G+ I+
Sbjct 806 GSRRGNDEIMARGTFANIRIVNKLMNGEVGPKTVHIPSGEKLSVFDAAMRYKSSGEDTII 865
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
LAG EYGSGSSRDWAAKGP LQGVKAVIA+SFERIHRSNLVGMGI+PL F G+ ADTL
Sbjct 866 LAGAEYGSGSSRDWAAKGPMLQGVKAVIAKSFERIHRSNLVGMGIIPLCFKSGEDADTLG 925
Query 120 LTGRERFNIAL--NGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLR 177
LTG ER+ I L + + PG V V+TD GKSF R DT+VE+ YF +GG L YV+R
Sbjct 926 LTGHERYTIHLPTDISEIRPGQDVTVTTDNGKSFTCTVRFDTEVELAYFNHGGILPYVIR 985
Query 178 NM 179
N+
Sbjct 986 NL 987
> pfa:PF13_0229 aconitase (EC:4.2.1.3); K01681 aconitate hydratase
1 [EC:4.2.1.3]
Length=909
Score = 226 bits (576), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 104/183 (56%), Positives = 136/183 (74%), Gaps = 3/183 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGNDEVM+RGTFANIRL+NK+CP GP +H+PTN+++ V + A +Y+ + +I++
Sbjct 722 GARRGNDEVMIRGTFANIRLINKLCPDKGPNTIHIPTNQLMSVYQAAMKYKQDNIDVIII 781
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYG GSSRDWAAKGP L GVKAVIAES+ERIHRSNL+GM +LPLQF+ QS +
Sbjct 782 AGKEYGCGSSRDWAAKGPNLLGVKAVIAESYERIHRSNLIGMSVLPLQFINNQSPQYYNM 841
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDC-GK--SFEAKCRIDTDVEVEYFRNGGALHYVLR 177
G E+F I LN G + ++V + GK F+ CRIDT++E YFRNGG L YVLR
Sbjct 842 DGTEKFTILLNDGNIKAQQTIKVQMNQKGKIIIFDVLCRIDTEIEERYFRNGGILKYVLR 901
Query 178 NML 180
+++
Sbjct 902 SLV 904
> dre:568448 aco1, wu:fo75b03; aconitase 1, soluble (EC:4.2.1.3);
K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=890
Score = 226 bits (575), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 105/179 (58%), Positives = 136/179 (75%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
GSRRGND VM RGTFANIRL NK P ++LPT E L V + AE+Y+ G P+++L
Sbjct 710 GSRRGNDAVMARGTFANIRLFNKFINKQSPTTIYLPTGETLDVFDAAEKYQQAGHPLLIL 769
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYGSGSSRDWAAKGP+L G+KAV+AES+ERIHRSNLVGMG++PL++L G SA++L L
Sbjct 770 AGKEYGSGSSRDWAAKGPFLLGIKAVLAESYERIHRSNLVGMGVIPLEYLPGDSAESLGL 829
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
+GRER+ + + L P V + D GK+F+A+ R DTDVE+ YF +GG L+Y++R M
Sbjct 830 SGRERYTVMI-PPLLKPRMTVDIKLDTGKTFQARMRFDTDVELTYFHHGGILNYMIRKM 887
> dre:652982 ireb2, im:7153062; iron-responsive element binding
protein 2
Length=896
Score = 221 bits (564), Expect = 8e-58, Method: Compositional matrix adjust.
Identities = 107/180 (59%), Positives = 136/180 (75%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGND VM RGTFA+I+L N++ GPK +H+PT + L V E AERY+ +G P+I+L
Sbjct 717 GARRGNDAVMTRGTFASIKLQNRLIGKTGPKTLHIPTGQTLDVFEAAERYQRDGVPLIIL 776
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGKEYGSGSSRDWAAKGPYL GV+AVIAESFE+IHR++LVGMGI PLQFL GQ+AD+L+L
Sbjct 777 AGKEYGSGSSRDWAAKGPYLLGVRAVIAESFEKIHRNHLVGMGIAPLQFLPGQNADSLEL 836
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
G+ERF I + L + V T GKSF + D++VE+FR+GG L YV R++L
Sbjct 837 CGKERFTIDI-PEELTARQQITVQTSTGKSFMVTALFENDMDVEFFRHGGILKYVARSLL 895
> ath:AT4G35830 aconitate hydratase, cytoplasmic / citrate hydro-lyase
/ aconitase (ACO); K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=795
Score = 220 bits (561), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 112/183 (61%), Positives = 135/183 (73%), Gaps = 3/183 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCP-SAGPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGNDE+M RGTFANIR+VNK GPK +H+PT E L V + A +YR EG+ I+
Sbjct 611 GSRRGNDEIMARGTFANIRIVNKHLKGEVGPKTVHIPTGEKLSVFDAAMKYRNEGRDTII 670
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
LAG EYGSGSSRDWAAKGP L GVKAVI++SFERIHRSNLVGMGI+PL F G+ A+TL
Sbjct 671 LAGAEYGSGSSRDWAAKGPMLLGVKAVISKSFERIHRSNLVGMGIIPLCFKAGEDAETLG 730
Query 120 LTGRERFNIAL--NGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLR 177
LTG+E + I L N + PG V V T+ GKSF R DT+VE+ YF +GG L YV+R
Sbjct 731 LTGQELYTIELPNNVSEIKPGQDVTVVTNNGKSFTCTLRFDTEVELAYFDHGGILQYVIR 790
Query 178 NML 180
N++
Sbjct 791 NLI 793
> ath:AT4G26970 aconitate hydratase/ copper ion binding (EC:4.2.1.3);
K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=995
Score = 213 bits (543), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 108/182 (59%), Positives = 131/182 (71%), Gaps = 3/182 (1%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCP-SAGPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGNDEVM RGTFANIR+VNK+ GP +H+PT E L V + A +Y+ Q I+
Sbjct 811 GSRRGNDEVMARGTFANIRIVNKLLKGEVGPNTVHIPTGEKLSVFDAASKYKTAEQDTII 870
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
LAG EYGSGSSRDWAAKGP L GVKAVIA+SFERIHRSNL GMGI+PL F G+ A+TL
Sbjct 871 LAGAEYGSGSSRDWAAKGPLLLGVKAVIAKSFERIHRSNLAGMGIIPLCFKAGEDAETLG 930
Query 120 LTGRERFNIALNG--GRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLR 177
LTG ER+ + L + PG V V+TD GKSF R DT+VE+ Y+ +GG L YV+R
Sbjct 931 LTGHERYTVHLPTKVSDIRPGQDVTVTTDSGKSFVCTLRFDTEVELAYYDHGGILPYVIR 990
Query 178 NM 179
++
Sbjct 991 SL 992
> hsa:3658 IREB2, ACO3, FLJ23381, IRP2, IRP2AD; iron-responsive
element binding protein 2
Length=963
Score = 203 bits (516), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 95/179 (53%), Positives = 127/179 (70%), Gaps = 1/179 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGND VM RGTFANI+L NK PK +H P+ + L V E AE Y+ EG P+I+L
Sbjct 785 GARRGNDAVMTRGTFANIKLFNKFIGKPAPKTIHFPSGQTLDVFEAAELYQKEGIPLIIL 844
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGK+YGSG+SRDWAAKGPYL GVKAV+AES+E+IH+ +L+G+GI PLQFL G++AD+L L
Sbjct 845 AGKKYGSGNSRDWAAKGPYLLGVKAVLAESYEKIHKDHLIGIGIAPLQFLPGENADSLGL 904
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNM 179
+GRE F++ L PG + + T GK F + DVE+ +++GG L++V R
Sbjct 905 SGRETFSLTFP-EELSPGITLNIQTSTGKVFSVIASFEDDVEITLYKHGGLLNFVARKF 962
> mmu:64602 Ireb2, D9Ertd85e, DKFZp564D1164, Irp2; iron responsive
element binding protein 2
Length=963
Score = 203 bits (516), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 95/180 (52%), Positives = 128/180 (71%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGND VM RGTFANI+L NK PK +H P+ + L V E AE Y+ EG P+I+L
Sbjct 785 GARRGNDAVMTRGTFANIKLFNKFIGKPAPKTIHFPSGQTLDVFEAAELYQKEGIPLIIL 844
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGK+YGSG+SRDWAAKGPYL GVKAV+AES+E+IH+ +L+G+GI PL+FL G++AD+L L
Sbjct 845 AGKKYGSGNSRDWAAKGPYLLGVKAVLAESYEKIHKDHLIGIGIAPLEFLPGENADSLGL 904
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
+GRE F+++ L PG + + T GK F DVE+ +++GG L++V R L
Sbjct 905 SGREVFSLSFP-EELFPGITLNIKTSTGKEFSVIASFANDVEITLYKHGGLLNFVARKFL 963
> eco:b1276 acnA, acn, ECK1271, JW1268; aconitate hydratase 1
(EC:4.2.1.3); K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=891
Score = 202 bits (515), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 102/185 (55%), Positives = 131/185 (70%), Gaps = 6/185 (3%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSA-GPKAMHLPTNEILPVSEVAERYRAEGQPMIV 59
GSRRGN EVM+RGTFANIR+ N+M P G HLP ++++ + + A RY+ E P+ V
Sbjct 707 GSRRGNHEVMMRGTFANIRIRNEMVPGVEGGMTRHLPDSDVVSIYDAAMRYKQEQTPLAV 766
Query 60 LAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLK 119
+AGKEYGSGSSRDWAAKGP L G++ VIAESFERIHRSNL+GMGILPL+F +G + TL
Sbjct 767 IAGKEYGSGSSRDWAAKGPRLLGIRVVIAESFERIHRSNLIGMGILPLEFPQGVTRKTLG 826
Query 120 LTGRERFNIA----LNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYV 175
LTG E+ +I L G VP ++ R + + +CRIDT E+ Y++N G LHYV
Sbjct 827 LTGEEKIDIGDLQNLQPGATVPVTLTR-ADGSQEVVPCRCRIDTATELTYYQNDGILHYV 885
Query 176 LRNML 180
+RNML
Sbjct 886 IRNML 890
> xla:414584 ireb2, MGC83131; iron-responsive element binding
protein 2
Length=955
Score = 196 bits (497), Expect = 5e-50, Method: Compositional matrix adjust.
Identities = 92/180 (51%), Positives = 129/180 (71%), Gaps = 1/180 (0%)
Query 1 GSRRGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVL 60
G+RRGND VM RGTFAN++L NK+ GPK +HLP+ + + V + AE Y+ P+I++
Sbjct 777 GARRGNDAVMTRGTFANMKLFNKLVGKTGPKTIHLPSGQTMDVFDAAELYQRSEIPLIII 836
Query 61 AGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKL 120
AGK+YG G+SRDWAAKGP+L GV+ VIAES+E+IH+ +LVGMGI PLQFL G++A+TL L
Sbjct 837 AGKKYGLGNSRDWAAKGPFLLGVRVVIAESYEKIHKDHLVGMGIAPLQFLSGENAETLGL 896
Query 121 TGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
+G+E+++++L L PG V + T+ GK F D + EV +++GG L YV R L
Sbjct 897 SGKEQYSLSLPVD-LTPGHKVEIKTNTGKIFHVIAAFDNEAEVTLYKHGGILSYVARKYL 955
> sce:YLR304C ACO1, GLU1; Aco1p (EC:4.2.1.3); K01681 aconitate
hydratase 1 [EC:4.2.1.3]
Length=778
Score = 76.6 bits (187), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 74/142 (52%), Gaps = 6/142 (4%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E V + A YR +G +V+ + +G GSSR+ AA P G A+I +SF RIH
Sbjct 634 TGEYKGVPDTARDYRDQGIKWVVIGDENFGEGSSREHAALEPRFLGGFAIITKSFARIHE 693
Query 97 SNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIV--RVSTDCGKSFEAK 154
+NL G+LPL F AD K+ +R +I L L PG V RV GK ++A
Sbjct 694 TNLKKQGLLPLNF--KNPADYDKINPDDRIDI-LGLAELAPGKPVTMRVHPKNGKPWDAV 750
Query 155 -CRIDTDVEVEYFRNGGALHYV 175
D ++E+F+ G AL+ +
Sbjct 751 LTHTFNDEQIEWFKYGSALNKI 772
> sce:YJL200C ACO2; Aco2p (EC:4.2.1.3); K01681 aconitate hydratase
1 [EC:4.2.1.3]
Length=789
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 76/138 (55%), Gaps = 17/138 (12%)
Query 49 RYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQ 108
+++++G+P V+A YG GS+R+ AA P G + ++ +SF RIH +NL G+LPL
Sbjct 651 KWKSDGRPWTVIAEHNYGEGSAREHAALSPRFLGGEILLVKSFARIHETNLKKQGVLPLT 710
Query 109 FLEGQSADTLKLTGR--ERFNIAL-------NGGRLVPGSIVRVSTDCGKSF--EAKCRI 157
F D + +G E N+ NGG + V+++ G+SF +AK +
Sbjct 711 FANESDYDKIS-SGDVLETLNLVDMIAKDGNNGGEID----VKITKPNGESFTIKAKHTM 765
Query 158 DTDVEVEYFRNGGALHYV 175
D ++++F+ G A++Y+
Sbjct 766 SKD-QIDFFKAGSAINYI 782
> hsa:50 ACO2, ACONM, MGC20605, MGC33908; aconitase 2, mitochondrial
(EC:4.2.1.3); K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=780
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 69/140 (49%), Gaps = 6/140 (4%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E PV + A Y+ G +V+ + YG GSSR+ AA P G +A+I +SF RIH
Sbjct 637 TQEFGPVPDTARYYKKHGIRWVVIGDENYGEGSSREHAALEPRHLGGRAIITKSFARIHE 696
Query 97 SNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCR 156
+NL G+LPL F + AD K+ ++ I PG ++ +
Sbjct 697 TNLKKQGLLPLTFAD--PADYNKIHPVDKLTIQ-GLKDFTPGKPLKCIIKHPNGTQETIL 753
Query 157 ID---TDVEVEYFRNGGALH 173
++ + ++E+FR G AL+
Sbjct 754 LNHTFNETQIEWFRAGSALN 773
> mmu:11429 Aco2, Aco-2, Aco3, D10Wsu183e; aconitase 2, mitochondrial
(EC:4.2.1.3); K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=780
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 68/146 (46%), Gaps = 18/146 (12%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E PV + A Y+ G +V+ + YG GSSR+ AA P G +A+I +SF RIH
Sbjct 637 TQEFGPVPDTARYYKKHGIRWVVIGDENYGEGSSREHAALEPRHLGGRAIITKSFARIHE 696
Query 97 SNLVGMGILPLQFLEG------QSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKS 150
+NL G+LPL F + D L + G + F PG ++
Sbjct 697 TNLKKQGLLPLTFADPSDYNKIHPVDKLTIQGLKDF---------APGKPLKCVIKHPNG 747
Query 151 FEAKCRID---TDVEVEYFRNGGALH 173
+ ++ + ++E+FR G AL+
Sbjct 748 TQETILLNHTFNETQIEWFRAGSALN 773
> dre:322670 aco2, cb1017, wu:fa10e03, wu:fb69g04, wu:fc20c11;
aconitase 2, mitochondrial (EC:4.2.1.3); K01681 aconitate hydratase
1 [EC:4.2.1.3]
Length=782
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 45/147 (30%), Positives = 76/147 (51%), Gaps = 8/147 (5%)
Query 31 KAMHLPTNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAES 90
K +L T E V +VA Y+ +V+ + YG GSSR+ AA P G +A+I +S
Sbjct 633 KLRNLLTGEYSGVPDVARHYKKNNISWVVVGDENYGEGSSREHAALEPRHLGGRAIIVKS 692
Query 91 FERIHRSNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPG----SIVRVSTD 146
F RIH +NL G+LPL F AD K+ ++ +I + PG ++++ +
Sbjct 693 FARIHETNLKKQGLLPLTF--SNPADYDKIRPDDKISI-IGLKSFAPGKPLKAVIKHTDG 749
Query 147 CGKSFEAKCRIDTDVEVEYFRNGGALH 173
++ E + + ++E+F+ G AL+
Sbjct 750 SSETIELNHTFN-ETQIEWFQAGSALN 775
> xla:444692 aco2, MGC84375; aconitase 2, mitochondrial (EC:4.2.1.3);
K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=782
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 46/134 (34%), Positives = 70/134 (52%), Gaps = 6/134 (4%)
Query 43 VSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGM 102
V + A Y+A G +V+ + YG GSSR+ AA P G +A+I +SF RIH +NL
Sbjct 645 VPDTARYYKAHGIKWVVIGDENYGEGSSREHAALEPRHLGGRAIITKSFARIHETNLKKQ 704
Query 103 GILPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVR-VSTDCGKSFEAKCRIDT-- 159
G+LPL F AD K+ ++ +A L PG V+ + T S E T
Sbjct 705 GLLPLTF--SDPADYDKIHPEDKITLA-GLKDLAPGKPVKCIITHQNGSQETIILNHTFN 761
Query 160 DVEVEYFRNGGALH 173
+ ++++F+ G AL+
Sbjct 762 ETQLQWFQAGSALN 775
> cel:F54H12.1 aco-2; ACOnitase family member (aco-2); K01681
aconitate hydratase 1 [EC:4.2.1.3]
Length=777
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 71/141 (50%), Gaps = 8/141 (5%)
Query 37 TNEILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHR 96
T E V A +Y+A+G + + + YG GSSR+ AA P G +A+I +SF RIH
Sbjct 631 TGEYGAVPATARKYKADGVRWVAIGDENYGEGSSREHAALEPRHLGGRAIIVKSFARIHE 690
Query 97 SNLVGMGILPLQFLEGQSADTLKLTGRERFNIALNG-GRLVPG---SIVRVSTDCGKSFE 152
+NL G+LPL F AD K+ + N+++ G PG + + T+ K
Sbjct 691 TNLKKQGMLPLTF--ANPADYDKIDPSD--NVSIVGLSSFAPGKPLTAIFKKTNGSKVEV 746
Query 153 AKCRIDTDVEVEYFRNGGALH 173
+ ++E+F+ G AL+
Sbjct 747 TLNHTFNEQQIEWFKAGSALN 767
> ath:AT5G54950 aconitate hydratase-related / citrate hydro-lyase-related
/ aconitase-related
Length=74
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 107 LQFLEGQSADTLKLTGRERFNIAL--NGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVE 164
+ F G+ A+TL LTG E + I L N + PG + V+TD KSF R+DT++ V
Sbjct 1 MAFKSGEDAETLGLTGHELYTIHLPSNINEIKPGQDITVTTDTAKSFVCTLRLDTEIWVL 60
Query 165 YF 166
F
Sbjct 61 CF 62
> xla:398139 aconitase; K01681 aconitate hydratase 1 [EC:4.2.1.3]
Length=856
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 43 VSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGM 102
V + A Y+A G +V+ + YG GSSR+ AA P G +A+I +SF RIH + + +
Sbjct 743 VPDTARYYKAHGIKWVVIGDENYGEGSSREHAALEPRHLGGRAIITKSFARIHPEDKITL 802
Query 103 GIL 105
L
Sbjct 803 AGL 805
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 43 VSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERI 94
V + A Y+A G +V+ + YG GSSR+ AA P G +A+I +SF RI
Sbjct 646 VPDTARYYKAHGIKWVVIGDENYGEGSSREHAALEPRHLGGRAIITKSFARI 697
> ath:AT2G43100 aconitase C-terminal domain-containing protein
(EC:4.2.1.33); K01704 3-isopropylmalate/(R)-2-methylmalate
dehydratase small subunit [EC:4.2.1.33 4.2.1.35]
Length=256
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 58 IVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMG-ILPLQ 108
I++ G+ +G GSSR+ A G KA++AES+ RI N V G + PL+
Sbjct 139 IIIGGENFGCGSSREHAPVCLGAAGAKAIVAESYARIFFRNSVATGEVFPLE 190
> sce:YGL009C LEU1; Isopropylmalate isomerase, catalyzes the second
step in the leucine biosynthesis pathway (EC:4.2.1.33);
K01702 3-isopropylmalate dehydratase [EC:4.2.1.33]
Length=779
Score = 37.0 bits (84), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 40/79 (50%), Gaps = 7/79 (8%)
Query 57 MIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSAD 116
++V+ G +G GSSR+ A G+K++IA S+ I +N G+LP++ + D
Sbjct 617 ILVVTGDNFGCGSSREHAPWALKDFGIKSIIAPSYGDIFYNNSFKNGLLPIRLDQQIIID 676
Query 117 TLKLTGRERFNIALNGGRL 135
L IA GG+L
Sbjct 677 KL-------IPIANKGGKL 688
> sce:YDR234W LYS4, LYS3; Homoaconitase, catalyzes the conversion
of homocitrate to homoisocitrate, which is a step in the
lysine biosynthesis pathway (EC:4.2.1.36); K01705 homoaconitate
hydratase [EC:4.2.1.36]
Length=693
Score = 35.8 bits (81), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 49 RYRAEGQP-MIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPL 107
+R + P IV++G +G+GSSR+ AA +G+ V++ SF I N + +L L
Sbjct 556 EFRTKVHPGDIVVSGFNFGTGSSREQAATALLAKGINLVVSGSFGNIFSRNSINNALLTL 615
Query 108 QF 109
+
Sbjct 616 EI 617
> eco:b0071 leuD, ECK0073, JW0070; 3-isopropylmalate dehydratase
small subunit (EC:4.2.1.33); K01704 3-isopropylmalate/(R)-2-methylmalate
dehydratase small subunit [EC:4.2.1.33 4.2.1.35]
Length=201
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 58 IVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADT 117
I+LA + +G GSSR+ A G K VIA SF I N +LP++ + + +
Sbjct 73 ILLARENFGCGSSREHAPWALTDYGFKVVIAPSFADIFYGNSFNNQLLPVKLSDAEVDEL 132
Query 118 LKLT 121
L
Sbjct 133 FALV 136
> cpv:cgd6_2300 Cut4/Apc1p/TSG24 family protein; meiotic check
point regulator and 26S proteasome regulatory complex; PC-rep
repeats
Length=2006
Score = 34.3 bits (77), Expect = 0.23, Method: Composition-based stats.
Identities = 19/44 (43%), Positives = 27/44 (61%), Gaps = 6/44 (13%)
Query 70 SRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMGILPLQFLEGQ 113
S+ WA+ YL+ K F+RI SNL+ GI+P +FLEG+
Sbjct 539 SKFWAS---YLEKNKK---SPFDRIEFSNLISKGIIPHEFLEGK 576
> eco:b0771 ybhJ, ECK0760, JW5103; predicted hydratase; K01681
aconitate hydratase 1 [EC:4.2.1.3]
Length=753
Score = 33.9 bits (76), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query 66 GSGSSRDWAAKGPYLQGVKAVIAESF-ERIHRSNLVGMGILPLQFLE 111
G GS+R+ AA + G A IAE + + +RSN++ G+LPLQ E
Sbjct 638 GDGSAREQAASCQRVIGGLANIAEEYATKRYRSNVINWGMLPLQMAE 684
> cel:F40G9.14 hypothetical protein
Length=261
Score = 32.3 bits (72), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 31/67 (46%), Gaps = 2/67 (2%)
Query 105 LPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAKCRIDTD--VE 162
L L+ L G+S DTL T + + R+ P V DC + F C+ + D VE
Sbjct 195 LILEVLNGKSKDTLSCTSCQLTYSSQTFPRIHPSCGHSVCEDCKEPFCKICKTEEDRVVE 254
Query 163 VEYFRNG 169
+ RNG
Sbjct 255 MILVRNG 261
> tgo:TGME49_016710 hypothetical protein
Length=593
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 130 LNGGRLVPGSIVRVSTDCGKSFEAKCRIDTDVEVEYFRNGGALHYVL 176
L+G L PG + V G + + + YF +GGA+H+ L
Sbjct 165 LSGANLFPGHVATVKAVLGAALSCAFVVPIAIRSVYFHSGGAVHHRL 211
> ath:AT5G57160 ATLIG4; ATLIG4; DNA ligase (ATP)/ protein binding;
K10777 DNA ligase 4 [EC:6.5.1.1]
Length=1219
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query 148 GKSFEAKCRIDTDVEVEYFRNGGALHYVLRNML 180
GK A+C+ D D ++ +NG +HY RN L
Sbjct 245 GKDVVAECKFDGD-RIQIHKNGTDIHYFSRNFL 276
> ath:AT3G58990 aconitase C-terminal domain-containing protein
(EC:4.2.1.33); K01704 3-isopropylmalate/(R)-2-methylmalate
dehydratase small subunit [EC:4.2.1.33 4.2.1.35]
Length=253
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 58 IVLAGKEYGSGSSRDWAAKGPYLQGVKAVIAESFERIHRSNLVGMG-ILPLQ 108
+++ G +G GSSR+ A G KAV+AES+ RI N V G I PL+
Sbjct 136 VIIGGDNFGCGSSREHAPVCLGAAGAKAVVAESYARIFFRNCVATGEIFPLE 187
> ath:AT4G33945 armadillo/beta-catenin repeat family protein
Length=464
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query 82 GVKAVIAESFERIHRSNLVGMGILPLQFLEGQSADTLKLTGRERFNI 128
GV+ + E+F+ + R N+ +G+ P + LE + TLKL G + F I
Sbjct 2 GVRTISQEAFDDLVRENVEDLGMDPSEALE-DALYTLKLQGVDLFGI 47
> cpv:cgd7_4390 NudC ortholog
Length=312
Score = 29.6 bits (65), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 21 VNKMCPSAGPKAMHLPTNEILPVSEVAERYRAEGQPMIVLAGK-EYGSG 68
V KM KAM LPT++ L E+ E+++A M K YGSG
Sbjct 263 VEKMMFDQRQKAMGLPTSDNLKQHELLEKFKAAHPEMDFSQAKINYGSG 311
> hsa:25992 SNED1, DKFZp586B2420, FLJ00133, SST3, Snep; sushi,
nidogen and EGF-like domains 1
Length=1413
Score = 29.6 bits (65), Expect = 6.0, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 1/50 (2%)
Query 105 LPLQFLEGQSADTLKLTGRERFNIALNGGRLVPGSIVRVSTDCGKSFEAK 154
L LQ E S D + G N NGG VPG+ S DCG F+ +
Sbjct 1292 LALQLPEHGSKDIGNVPGNCSENPCQNGGTCVPGADAH-SCDCGPGFKGR 1340
> ath:AT5G04500 glycosyltransferase family protein 47
Length=765
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 24/37 (64%), Gaps = 3/37 (8%)
Query 129 ALNGGR--LVPGSIVRVSTDCGKSFEAKCRIDTDVEV 163
A NGGR L GS+ RV DCG+++ + R+ + +EV
Sbjct 298 ARNGGRAFLYDGSLYRVGQDCGENYGKRIRV-SKIEV 333
> mmu:54667 Atp8b2, Id; ATPase, class I, type 8B, member 2 (EC:3.6.3.1);
K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1214
Score = 29.3 bits (64), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 31/72 (43%), Gaps = 1/72 (1%)
Query 4 RGNDEVMVRGTFANIRLVNKMCPSAGPKAMHLPTNEIL-PVSEVAERYRAEGQPMIVLAG 62
R V+VR IRL K + +H PT E+L ++ Y +G +VLA
Sbjct 541 RKRMSVIVRNPEGKIRLYCKGADTILLDRLHPPTQELLSSTTDHLNEYAGDGLRTLVLAY 600
Query 63 KEYGSGSSRDWA 74
K+ +WA
Sbjct 601 KDLDEEYYEEWA 612
> hsa:100506627 DCDC5, DKFZp779D068, FLJ44078, FLJ46154, KIAA1493,
MGC190499, MGC190811; doublecortin domain containing 5
Length=890
Score = 29.3 bits (64), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 5/60 (8%)
Query 31 KAMHLPTNE--ILPVSEVAERYRAEGQPMIVLAGKEYGSGSSRDWAAKGPYLQGVKAVIA 88
+ HL +N +L VS R G P+IV K Y +G++ K Y++ +KA++A
Sbjct 325 RTFHLVSNPDLVLAVSMTKTRNEVCGYPVIVQKYKPYNNGAANQ---KWHYMKNIKALVA 381
Lambda K H
0.319 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4859948100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40