bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1169_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
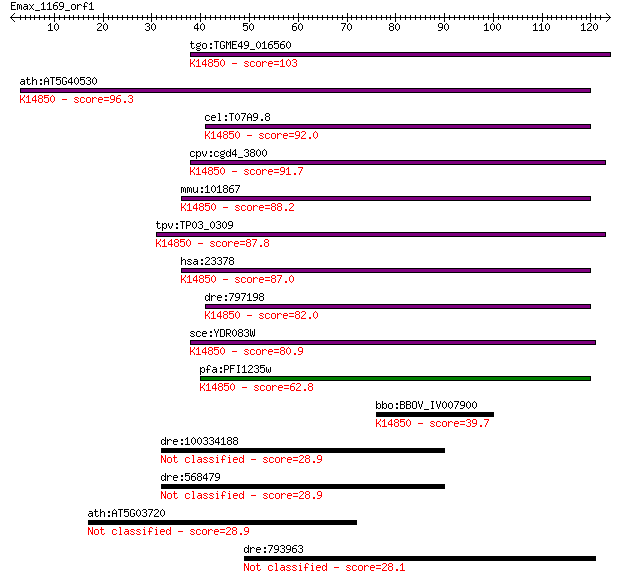
tgo:TGME49_016560 hypothetical protein ; K14850 ribosomal RNA-... 103 1e-22
ath:AT5G40530 hypothetical protein; K14850 ribosomal RNA-proce... 96.3 2e-20
cel:T07A9.8 hypothetical protein; K14850 ribosomal RNA-process... 92.0 4e-19
cpv:cgd4_3800 Rrp8p like methyltransferase involved in rRNA pr... 91.7 5e-19
mmu:101867 Rrp8, 1500003O22Rik, 2900001K19Rik, AW538116; ribos... 88.2 6e-18
tpv:TP03_0309 hypothetical protein; K14850 ribosomal RNA-proce... 87.8 8e-18
hsa:23378 RRP8, KIAA0409; ribosomal RNA processing 8, methyltr... 87.0 1e-17
dre:797198 fk33d07, si:ch211-152b13.1; wu:fk33d07; K14850 ribo... 82.0 4e-16
sce:YDR083W RRP8; Nucleolar protein involved in rRNA processin... 80.9 9e-16
pfa:PFI1235w methyltransferase, putative; K14850 ribosomal RNA... 62.8 3e-10
bbo:BBOV_IV007900 23.m06355; hypothetical protein; K14850 ribo... 39.7 0.002
dre:100334188 T-box 3 protein-like 28.9 4.0
dre:568479 T-box 3 protein-like 28.9 4.0
ath:AT5G03720 AT-HSFA3; DNA binding / transcription factor 28.9 4.2
dre:793963 G protein-coupled receptor 112-like 28.1 7.5
> tgo:TGME49_016560 hypothetical protein ; K14850 ribosomal RNA-processing
protein 8 [EC:2.1.1.-]
Length=610
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 42/86 (48%), Positives = 55/86 (63%), Gaps = 0/86 (0%)
Query 38 VIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVAT 97
++ +LQG RFRSLN+ LYT G +A + K+P LF YH GYR QV +WP NPL +
Sbjct 270 LLQKLQGSRFRSLNQCLYTSTGDQALAAFTKDPSLFHAYHEGYRLQVAQWPSNPLTHIKA 329
Query 98 WLSKKPKEWVVGDFGCGEAALALRFP 123
W+ P W++ D GCG+A LA FP
Sbjct 330 WVRTLPASWIIADLGCGDADLAKSFP 355
> ath:AT5G40530 hypothetical protein; K14850 ribosomal RNA-processing
protein 8 [EC:2.1.1.-]
Length=287
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 50/128 (39%), Positives = 71/128 (55%), Gaps = 11/128 (8%)
Query 3 PSADEKRETMANLQQQKERQLLLVKKFSGRVAA-------AGVID----RLQGGRFRSLN 51
PS +E ET Q +K+ Q + G +A + +D RL GG+FR LN
Sbjct 22 PSKEEPIETTPKNQNEKKNQRDTKNQQHGGSSAPSKRPKPSNFLDALRERLSGGQFRMLN 81
Query 52 EYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWLSKKPKEWVVGDF 111
E LYT G EA ++++PQ+FD+YHTGY+ Q+ WP P++ + WL VV DF
Sbjct 82 EKLYTCSGKEALDYFKEDPQMFDMYHTGYQQQMSNWPELPVNSIINWLLSNSSSLVVADF 141
Query 112 GCGEAALA 119
GCG+A +A
Sbjct 142 GCGDARIA 149
> cel:T07A9.8 hypothetical protein; K14850 ribosomal RNA-processing
protein 8 [EC:2.1.1.-]
Length=343
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 41/79 (51%), Positives = 53/79 (67%), Gaps = 0/79 (0%)
Query 41 RLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWLS 100
RL GRFR LNE LYT GSEAF ++++P FD+YH G+ QV++WP +PL + WL
Sbjct 133 RLDAGRFRFLNEKLYTCTGSEAFDFFKEDPTAFDLYHKGFADQVKKWPNHPLREIIRWLQ 192
Query 101 KKPKEWVVGDFGCGEAALA 119
KP + V D GCGEA +A
Sbjct 193 SKPDQQSVFDLGCGEAKIA 211
> cpv:cgd4_3800 Rrp8p like methyltransferase involved in rRNA
processing ; K14850 ribosomal RNA-processing protein 8 [EC:2.1.1.-]
Length=369
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 39/85 (45%), Positives = 53/85 (62%), Gaps = 0/85 (0%)
Query 38 VIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVAT 97
V RLQG FR +NE+LYT +AF +Y K+ +F+ YH GY Q R WP++PLD +
Sbjct 83 VSTRLQGSLFRKINEFLYTSDSEKAFNEYIKDGNMFENYHKGYEIQKRSWPIDPLDNIIN 142
Query 98 WLSKKPKEWVVGDFGCGEAALALRF 122
++SK V+GDFGCG A + F
Sbjct 143 YISKNKHLKVIGDFGCGTAKIGQTF 167
> mmu:101867 Rrp8, 1500003O22Rik, 2900001K19Rik, AW538116; ribosomal
RNA processing 8, methyltransferase, homolog (yeast);
K14850 ribosomal RNA-processing protein 8 [EC:2.1.1.-]
Length=457
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 56/84 (66%), Gaps = 0/84 (0%)
Query 36 AGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCV 95
A + RL G RFR LNE LY+ S A +Q++P+ F +YH G++ QV++WPL+P+D +
Sbjct 241 ARMTQRLDGARFRYLNEQLYSGPSSAARRLFQEDPEAFLLYHRGFQRQVKKWPLHPVDRI 300
Query 96 ATWLSKKPKEWVVGDFGCGEAALA 119
A L +KP VV DFGCG+ LA
Sbjct 301 AKDLRQKPASLVVADFGCGDCRLA 324
> tpv:TP03_0309 hypothetical protein; K14850 ribosomal RNA-processing
protein 8 [EC:2.1.1.-]
Length=236
Score = 87.8 bits (216), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 54/92 (58%), Gaps = 0/92 (0%)
Query 31 GRVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLN 90
G+ + RL G RFR +NE LY K +F + +P+L+ YH GYR QV WP N
Sbjct 12 GKRGLEEIRSRLSGSRFRCINEKLYKCKSDISFTMFNSDPKLYSAYHEGYRNQVLTWPYN 71
Query 91 PLDCVATWLSKKPKEWVVGDFGCGEAALALRF 122
P+D V WL ++ + +GDFGCG+A +A F
Sbjct 72 PVDKVIQWLKQRQELVNIGDFGCGDALIAKTF 103
> hsa:23378 RRP8, KIAA0409; ribosomal RNA processing 8, methyltransferase,
homolog (yeast); K14850 ribosomal RNA-processing
protein 8 [EC:2.1.1.-]
Length=456
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 56/84 (66%), Gaps = 0/84 (0%)
Query 36 AGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCV 95
A + RL G RFR LNE LY+ S A +Q++P+ F +YH G+++QV++WPL P+D +
Sbjct 240 ARMAQRLDGARFRYLNEQLYSGPSSAAQRLFQEDPEAFLLYHRGFQSQVKKWPLQPVDRI 299
Query 96 ATWLSKKPKEWVVGDFGCGEAALA 119
A L ++P VV DFGCG+ LA
Sbjct 300 ARDLRQRPASLVVADFGCGDCRLA 323
> dre:797198 fk33d07, si:ch211-152b13.1; wu:fk33d07; K14850 ribosomal
RNA-processing protein 8 [EC:2.1.1.-]
Length=533
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 34/79 (43%), Positives = 50/79 (63%), Gaps = 0/79 (0%)
Query 41 RLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWLS 100
+L+ RFR +NE LYT + A +Q++P IYH GY QV+ WP NP+D + +++
Sbjct 324 QLEAARFRFINEQLYTSTSAAAKRMFQQDPDAITIYHKGYTTQVQHWPTNPVDSIISYIC 383
Query 101 KKPKEWVVGDFGCGEAALA 119
+KP VV DFGCG+ +A
Sbjct 384 QKPASLVVADFGCGDCKIA 402
> sce:YDR083W RRP8; Nucleolar protein involved in rRNA processing,
pre-rRNA cleavage at site A2; also involved in telomere
maintenance; mutation is synthetically lethal with a gar1 mutation;
K14850 ribosomal RNA-processing protein 8 [EC:2.1.1.-]
Length=392
Score = 80.9 bits (198), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 41/99 (41%), Positives = 54/99 (54%), Gaps = 16/99 (16%)
Query 38 VIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVAT 97
++ +L G RFR +NE LYT EA +++PQLFD YH G+R+QV+ WP NP+D
Sbjct 117 MMAKLTGSRFRWINEQLYTISSDEALKLIKEQPQLFDEYHDGFRSQVQAWPENPVDVFVD 176
Query 98 WLS----------------KKPKEWVVGDFGCGEAALAL 120
+ K KE V+ D GCGEA LAL
Sbjct 177 QIRYRCMKPVNAPGGLPGLKDSKEIVIADMGCGEAQLAL 215
> pfa:PFI1235w methyltransferase, putative; K14850 ribosomal RNA-processing
protein 8 [EC:2.1.1.-]
Length=413
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 30/81 (37%), Positives = 42/81 (51%), Gaps = 1/81 (1%)
Query 40 DRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWL 99
D + FR +NEY+YT K + +F+IYH GY+ Q +WP NP+ + L
Sbjct 194 DIVNSSLFRYINEYMYTNNSEVVQNKLNQTKNVFNIYHQGYKNQKNKWPHNPVSIIIKHL 253
Query 100 SKK-PKEWVVGDFGCGEAALA 119
K K + D GCGEA +A
Sbjct 254 KKYFNKNNKIADLGCGEAEIA 274
> bbo:BBOV_IV007900 23.m06355; hypothetical protein; K14850 ribosomal
RNA-processing protein 8 [EC:2.1.1.-]
Length=128
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 76 YHTGYRAQVRRWPLNPLDCVATWL 99
YH GYR QV +WP++PL+ + WL
Sbjct 13 YHEGYREQVEKWPIDPLNKILVWL 36
> dre:100334188 T-box 3 protein-like
Length=585
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 32 RVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPL 89
R+ + GV+ QG F +Y+ T GS + YQ Q F ++ YR Q R +P+
Sbjct 477 RMTSQGVLVSAQGNIFPYPQDYMTTSAGSSSAASYQVHRQPF--FNISYRPQCRPYPV 532
> dre:568479 T-box 3 protein-like
Length=585
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 32 RVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPL 89
R+ + GV+ QG F +Y+ T GS + YQ Q F ++ YR Q R +P+
Sbjct 477 RMTSQGVLVSAQGNIFPYPQDYMTTSAGSSSAASYQVHRQPF--FNISYRPQCRPYPV 532
> ath:AT5G03720 AT-HSFA3; DNA binding / transcription factor
Length=412
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 32/57 (56%), Gaps = 6/57 (10%)
Query 17 QQKERQLL--LVKKFSGRVAAAGVIDRLQGGRFRSLNEYLYTKKGSEAFVKYQKEPQ 71
+Q+++QLL L K F R G ++RL+ + + L +K + F+K+ ++PQ
Sbjct 211 EQRQKQLLSFLAKLFQNR----GFLERLKNFKGKEKGGALGLEKARKKFIKHHQQPQ 263
> dre:793963 G protein-coupled receptor 112-like
Length=806
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 33/75 (44%), Gaps = 14/75 (18%)
Query 49 SLNEYLYTKKGSEAFVKYQKEPQLFDIYHTGYRAQVRRWPLNPLDCVATWLSKKPKEWV- 107
+LN Y+ + + A V + P + + H + + + P+NP D EWV
Sbjct 431 TLNSYVVSASVTNATVSELEIPVMVTLRHLQNKEDISKTPINPKD-----------EWVL 479
Query 108 --VGDFGCGEAALAL 120
+ GCG +++ L
Sbjct 480 TIISYVGCGISSIFL 494
Lambda K H
0.320 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40