bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
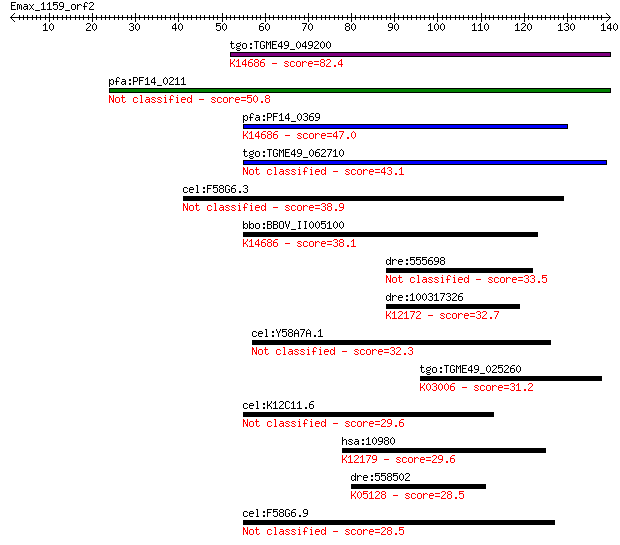
Query= Emax_1159_orf2
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_049200 ctr copper transporter domain-containing pro... 82.4 3e-16
pfa:PF14_0211 Ctr copper transporter domain containing protein... 50.8 1e-06
pfa:PF14_0369 copper transporter putative; K14686 solute carri... 47.0 2e-05
tgo:TGME49_062710 ctr copper transporter domain-containing pro... 43.1 3e-04
cel:F58G6.3 hypothetical protein 38.9 0.005
bbo:BBOV_II005100 18.m09998; hypothetical protein; K14686 solu... 38.1 0.007
dre:555698 peptidylprolyl isomerase F-like 33.5 0.21
dre:100317326 si:ch1073-55a19.2; K12172 E3 SUMO-protein ligase... 32.7 0.37
cel:Y58A7A.1 hypothetical protein 32.3 0.41
tgo:TGME49_025260 DNA-directed RNA polymerase II largest subun... 31.2 0.95
cel:K12C11.6 hypothetical protein 29.6 3.0
hsa:10980 COPS6, CSN6, MOV34-34KD; COP9 constitutive photomorp... 29.6 3.1
dre:558502 ryk, wu:fb37b12, wu:fi05b06, zgc:158381; receptor-l... 28.5 6.3
cel:F58G6.9 hypothetical protein 28.5 7.0
> tgo:TGME49_049200 ctr copper transporter domain-containing protein
; K14686 solute carrier family 31 (copper transporter),
member 1
Length=235
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 52/88 (59%), Gaps = 0/88 (0%)
Query 52 ALPLPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLRRLSEAHLKMS 111
+PLPM FE ST V+ LF W TT Q+ CV + GFI + LKV+RR E L
Sbjct 71 GMPLPMAFEVSTRVIYLFEDWPTETTTQFAGACVATCILGFICVILKVVRRYVEKSLVSQ 130
Query 112 EQKGKPTLLLGSLPVYQNAIRGSVAFLN 139
E GK L+ GS P+Y N++R VAF+N
Sbjct 131 ENVGKTKLIFGSFPLYSNSVRFLVAFVN 158
> pfa:PF14_0211 Ctr copper transporter domain containing protein,
putative
Length=160
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 58/116 (50%), Gaps = 2/116 (1%)
Query 24 AFCTAALFPLISTASIRVNTDCAMLDKCALPLPMWFEASTEVLLLFSWWNGRTTAQYIVC 83
+ T A+F L T ++ C + + LPM+F + + +LF + + Q+I C
Sbjct 10 KYYTLAIFCLTFTFDFVSSSCCHSKNDDGVMLPMYFSNNENIKMLFDIFQVKNRYQFIFC 69
Query 84 CVCCIVFGFISIALKVLRRLSEAHLKMSEQKGKPTLLLGSLPVYQNAIRGSVAFLN 139
+ CI+ GF S+ +KVL++ +++ G + + ++ QN G ++FL+
Sbjct 70 NILCIIMGFFSVYIKVLKKRLHHNVQKVADGGDGSYV--NMSPCQNVNYGFLSFLH 123
> pfa:PF14_0369 copper transporter putative; K14686 solute carrier
family 31 (copper transporter), member 1
Length=235
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Query 55 LPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLRRLSEAHLKMSEQK 114
+PM F+ +T ++LF+ W ++ Y + V C FG IS+ KV+R E L +E
Sbjct 91 MPMSFQLTTHTIILFNKWETKSALSYYISLVLCFFFGIISVGFKVVRLNVEQALPKTED- 149
Query 115 GKPTLLLGSLPVYQN 129
T + SL +++N
Sbjct 150 ---TNIFKSLVLFKN 161
> tgo:TGME49_062710 ctr copper transporter domain-containing protein
Length=375
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 50/108 (46%), Gaps = 24/108 (22%)
Query 55 LPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLR-----------RL 103
+PM F+ S ++LF W QY++ + C+V G +S+ LKVLR R
Sbjct 206 MPMSFQNSLHTVILFHSWETLERWQYVLSLLTCVVLGMLSVVLKVLRLRLEFFLAKRDRA 265
Query 104 SEAHL---KMSEQKGKP----------TLLLGSLPVYQNAIRGSVAFL 138
+E K+ E++G+ L G+ P+ QN+ R AF+
Sbjct 266 AEDAQRVEKLKEKEGQSSAASPSSAIVERLCGNFPLKQNSWRMLEAFV 313
> cel:F58G6.3 hypothetical protein
Length=134
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 43/89 (48%), Gaps = 1/89 (1%)
Query 41 VNTDCAMLDKCALP-LPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKV 99
+N + +D P + MWF + +LFS WN + + + C+ + G I A+K
Sbjct 1 MNHNSMDMDMNQGPFMWMWFHTKPQDTVLFSTWNITSAGKMVWACILVAIAGIILEAIKY 60
Query 100 LRRLSEAHLKMSEQKGKPTLLLGSLPVYQ 128
RRL + S+++ + LL ++ +Q
Sbjct 61 NRRLIQKRQSPSKKESYISRLLSTMHFFQ 89
> bbo:BBOV_II005100 18.m09998; hypothetical protein; K14686 solute
carrier family 31 (copper transporter), member 1
Length=297
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 30/68 (44%), Gaps = 0/68 (0%)
Query 55 LPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLRRLSEAHLKMSEQK 114
+PM+FE + + ++LF +W T QY V V +++ LK R L
Sbjct 163 MPMYFENTVKTVILFHFWKTTTGTQYAVSLFFIFVLSLMTVFLKAFRNKLNCALLQRPNG 222
Query 115 GKPTLLLG 122
PT+ G
Sbjct 223 YHPTVKYG 230
> dre:555698 peptidylprolyl isomerase F-like
Length=336
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 21/34 (61%), Gaps = 4/34 (11%)
Query 88 IVFGFISIALKVLRRLSEAHLKMSEQKGKPTLLL 121
+VFGFI + VLRR+ E M ++GKPT +
Sbjct 299 VVFGFIREGMNVLRRIGE----MGSKEGKPTHRI 328
> dre:100317326 si:ch1073-55a19.2; K12172 E3 SUMO-protein ligase
RanBP2
Length=2950
Score = 32.7 bits (73), Expect = 0.37, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 22/31 (70%), Gaps = 4/31 (12%)
Query 88 IVFGFISIALKVLRRLSEAHLKMSEQKGKPT 118
+ FGF++ ++VLRRL+E M ++GKPT
Sbjct 2913 VAFGFVTDGMQVLRRLAE----MGTKEGKPT 2939
> cel:Y58A7A.1 hypothetical protein
Length=130
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 17/69 (24%), Positives = 27/69 (39%), Gaps = 0/69 (0%)
Query 57 MWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLRRLSEAHLKMSEQKGK 116
M F TE +LF +W T V C ++ F+ L+ R +A ++ +
Sbjct 1 MSFHFGTEETILFDFWKTETAVGIAVACFITVLLAFLMETLRFFRDYRKAQTQLHQPPIS 60
Query 117 PTLLLGSLP 125
P L P
Sbjct 61 PEDRLKRSP 69
> tgo:TGME49_025260 DNA-directed RNA polymerase II largest subunit,
putative (EC:2.7.7.6 3.5.1.16); K03006 DNA-directed RNA
polymerase II subunit RPB1 [EC:2.7.7.6]
Length=1892
Score = 31.2 bits (69), Expect = 0.95, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 4/46 (8%)
Query 96 ALKVLRRLSEAHLKM----SEQKGKPTLLLGSLPVYQNAIRGSVAF 137
A VLRR+SE LKM E+ + +L +LP+ A+R SV +
Sbjct 216 AYAVLRRISEEDLKMMGFDPERAHPASFILSTLPIPPLAVRPSVQY 261
> cel:K12C11.6 hypothetical protein
Length=132
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 27/59 (45%), Gaps = 1/59 (1%)
Query 55 LPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLR-RLSEAHLKMSE 112
+ MWF T+ +LF WN T + C +V G + +K LR ++ + H E
Sbjct 9 MHMWFHTKTQDTVLFKTWNVTDTPTMVWVCCIIVVAGILLELIKFLRWKIEKWHKNRDE 67
> hsa:10980 COPS6, CSN6, MOV34-34KD; COP9 constitutive photomorphogenic
homolog subunit 6 (Arabidopsis); K12179 COP9 signalosome
complex subunit 6
Length=327
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 29/49 (59%), Gaps = 2/49 (4%)
Query 78 AQYIVCCVCCIVFGFISIALK--VLRRLSEAHLKMSEQKGKPTLLLGSL 124
A + + C V G +S+AL V+ +S+ ++M Q+G+P ++G+L
Sbjct 24 AAVVPSVMACGVTGSVSVALHPLVILNISDHWIRMRSQEGRPVQVIGAL 72
> dre:558502 ryk, wu:fb37b12, wu:fi05b06, zgc:158381; receptor-like
tyrosine kinase (EC:2.7.10.1); K05128 RYK receptor-like
tyrosine kinase [EC:2.7.10.1]
Length=604
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 80 YIVCCVCCIVFGFISIALKVLRRLSEAHLKM 110
YI CVCCIV ++I L VL S ++M
Sbjct 227 YISVCVCCIVIFLVAIILAVLHLHSMKRVEM 257
> cel:F58G6.9 hypothetical protein
Length=156
Score = 28.5 bits (62), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 15/72 (20%), Positives = 27/72 (37%), Gaps = 0/72 (0%)
Query 55 LPMWFEASTEVLLLFSWWNGRTTAQYIVCCVCCIVFGFISIALKVLRRLSEAHLKMSEQK 114
+ MW+ E +LF W + C G + ALK R +E +K+ ++
Sbjct 21 MWMWYHVDVEDTVLFKSWTVFDAGTMVWTCFVVAAAGILLEALKYARWATEERMKIDQEN 80
Query 115 GKPTLLLGSLPV 126
G + +
Sbjct 81 VDSKTKYGGIKI 92
Lambda K H
0.330 0.140 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40