bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1149_orf4
Length=279
Score E
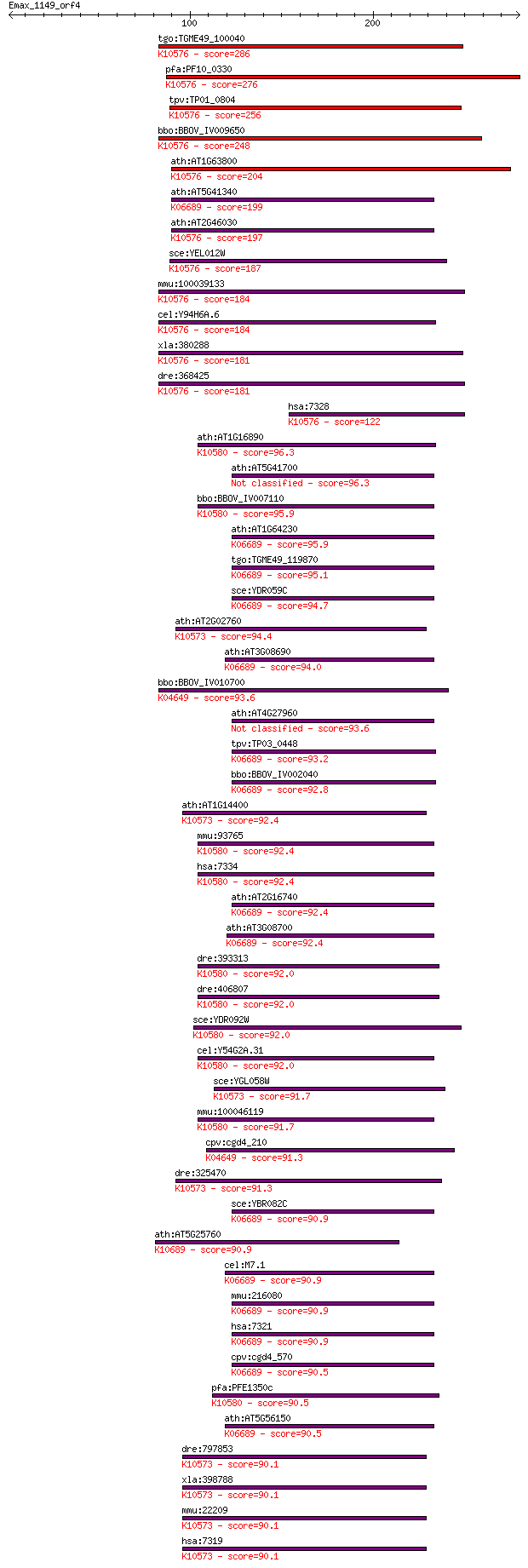
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_100040 ubiquitin-conjugating enzyme E2, putative (E... 286 7e-77
pfa:PF10_0330 ubiquitin conjugating enzyme, putative; K10576 u... 276 6e-74
tpv:TP01_0804 ubiquitin-protein ligase; K10576 ubiquitin-conju... 256 7e-68
bbo:BBOV_IV009650 23.m05904; ubiquitin-conjugating enzyme subu... 248 3e-65
ath:AT1G63800 UBC5; UBC5 (ubiquitin-conjugating enzyme 5); ubi... 204 2e-52
ath:AT5G41340 UBC4; UBC4 (UBIQUITIN CONJUGATING ENZYME 4); ubi... 199 7e-51
ath:AT2G46030 UBC6; UBC6 (ubiquitin-conjugating enzyme 6); ubi... 197 2e-50
sce:YEL012W UBC8, GID3; Ubc8p (EC:6.3.2.19); K10576 ubiquitin-... 187 5e-47
mmu:100039133 Gm2058; predicted gene 2058; K10576 ubiquitin-co... 184 2e-46
cel:Y94H6A.6 ubc-8; UBiquitin Conjugating enzyme family member... 184 3e-46
xla:380288 ube2h, MGC53494; ubiquitin-conjugating enzyme E2H (... 181 2e-45
dre:368425 ube2h, fc07a05, fi24c01, si:bz3c13.2, wu:fc07a05, w... 181 3e-45
hsa:7328 UBE2H, E2-20K, UBC8, UBCH, UBCH2; ubiquitin-conjugati... 122 1e-27
ath:AT1G16890 UBC36; ubiquitin-conjugating enzyme, putative; K... 96.3 1e-19
ath:AT5G41700 UBC8; UBC8 (UBIQUITIN CONJUGATING ENZYME 8); pro... 96.3 1e-19
bbo:BBOV_IV007110 23.m05993; ubiquitin-conjugating enzyme E2; ... 95.9 1e-19
ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative; K... 95.9 2e-19
tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containi... 95.1 2e-19
sce:YDR059C UBC5; Ubc5p (EC:6.3.2.19); K06689 ubiquitin-conjug... 94.7 3e-19
ath:AT2G02760 ATUBC2; ATUBC2 (UBIQUITING-CONJUGATING ENZYME 2)... 94.4 4e-19
ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11); ... 94.0 5e-19
bbo:BBOV_IV010700 23.m05926; ubiquitin-conjugating enzyme; K04... 93.6 7e-19
ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubi... 93.6 8e-19
tpv:TP03_0448 ubiquitin-conjugating enzyme (EC:6.3.2.19); K066... 93.2 9e-19
bbo:BBOV_IV002040 21.m02810; ubiquitin-conjugating enzyme E2 (... 92.8 1e-18
ath:AT1G14400 UBC1; UBC1 (UBIQUITIN CARRIER PROTEIN 1); ubiqui... 92.4 1e-18
mmu:93765 Ube2n, 1500026J17Rik, AL022654, BB101821, UBC13; ubi... 92.4 2e-18
hsa:7334 UBE2N, MGC131857, MGC8489, UBC13, UbcH-ben; ubiquitin... 92.4 2e-18
ath:AT2G16740 UBC29; UBC29 (ubiquitin-conjugating enzyme 29); ... 92.4 2e-18
ath:AT3G08700 UBC12; UBC12 (ubiquitin-conjugating enzyme 12); ... 92.4 2e-18
dre:393313 ube2nb, MGC63901, ube2nl, zgc:63901; ubiquitin-conj... 92.0 2e-18
dre:406807 ube2na, ube2n, wu:fb11b03, wu:fc08b06, zgc:55726; u... 92.0 2e-18
sce:YDR092W UBC13; Ubc13p (EC:6.3.2.19); K10580 ubiquitin-conj... 92.0 2e-18
cel:Y54G2A.31 ubc-13; UBiquitin Conjugating enzyme family memb... 92.0 2e-18
sce:YGL058W RAD6, PSO8, UBC2; Rad6p (EC:6.3.2.19); K10573 ubiq... 91.7 3e-18
mmu:100046119 Mcg1038069; ubiquitin-conjugating enzyme E2 N-li... 91.7 3e-18
cpv:cgd4_210 Ubc1p like ubiquitin-conjugating enzyme E2 fused ... 91.3 3e-18
dre:325470 fc79h10, wu:fc79h10; zgc:55512 (EC:6.3.2.19); K1057... 91.3 4e-18
sce:YBR082C UBC4; Ubc4p (EC:6.3.2.19); K06689 ubiquitin-conjug... 90.9 4e-18
ath:AT5G25760 PEX4; PEX4 (PEROXIN4); protein binding / ubiquit... 90.9 5e-18
cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquit... 90.9 5e-18
mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzy... 90.9 5e-18
hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiqui... 90.9 5e-18
cpv:cgd4_570 ubiquitin-conjugating enzyme ; K06689 ubiquitin-c... 90.5 6e-18
pfa:PFE1350c ubiquitin conjugating enzyme, putative; K10580 ub... 90.5 6e-18
ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30); ... 90.5 7e-18
dre:797853 ube2a, MGC64109, mp:zf637-2-000771, si:bz46j2.4, wu... 90.1 7e-18
xla:398788 ube2a, MGC68540; ubiquitin-conjugating enzyme E2A (... 90.1 7e-18
mmu:22209 Ube2a, HHR6A, HR6A, Mhr6a; ubiquitin-conjugating enz... 90.1 7e-18
hsa:7319 UBE2A, HHR6A, RAD6A, UBC2; ubiquitin-conjugating enzy... 90.1 7e-18
> tgo:TGME49_100040 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10576 ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=240
Score = 286 bits (731), Expect = 7e-77, Method: Compositional matrix adjust.
Identities = 129/166 (77%), Positives = 152/166 (91%), Gaps = 0/166 (0%)
Query 83 MTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 142
++ Q+SS+RKQCDF KL+MAG+DLELNN +TQDF+VVFHGPK T+YEGGVW+VHVTLPD
Sbjct 9 ISNHQVSSNRKQCDFTKLMMAGFDLELNNDSTQDFHVVFHGPKGTVYEGGVWKVHVTLPD 68
Query 143 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 202
DYPF+SPSIGF+NK+LHPNVDEASGSVCLDVINQTWTPLYSLVNVF+VFLPQLLTYPNP
Sbjct 69 DYPFASPSIGFMNKMLHPNVDEASGSVCLDVINQTWTPLYSLVNVFEVFLPQLLTYPNPT 128
Query 203 DPLNSEAASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEE 248
DPLNSEAASL++RDK YE+KV+E+V YASR+ WE+EQ KK+ +
Sbjct 129 DPLNSEAASLMSRDKRLYEEKVREYVRLYASREEWEKEQREKKEAQ 174
> pfa:PF10_0330 ubiquitin conjugating enzyme, putative; K10576
ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=191
Score = 276 bits (706), Expect = 6e-74, Method: Compositional matrix adjust.
Identities = 133/193 (68%), Positives = 155/193 (80%), Gaps = 5/193 (2%)
Query 87 QISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPF 146
Q S +RKQCDF KL+MAGYDLELNN +TQDF+V+FHGP T YEGG+W+VHVTLPDDYPF
Sbjct 4 QTSLTRKQCDFTKLIMAGYDLELNNGSTQDFDVMFHGPNGTAYEGGIWKVHVTLPDDYPF 63
Query 147 SSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLN 206
+SPSIGF+NK+LHPNVDEASGSVCLDVINQTWTPLYSLVNVF+VFLPQLLTYPNP DPLN
Sbjct 64 ASPSIGFMNKLLHPNVDEASGSVCLDVINQTWTPLYSLVNVFEVFLPQLLTYPNPSDPLN 123
Query 207 SEAASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEEAQERRREVEEPQLRELSR 266
S+AASLL +DK YE+KVKE+V YAS+D WE+++ KK V ELS
Sbjct 124 SDAASLLMKDKNIYEEKVKEYVKLYASKDLWEQQKKEKKPSNINGNISPV-----SELSY 178
Query 267 VDDEADGADIDSL 279
VD + D+D+L
Sbjct 179 VDQDVQDIDLDNL 191
> tpv:TP01_0804 ubiquitin-protein ligase; K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=189
Score = 256 bits (654), Expect = 7e-68, Method: Compositional matrix adjust.
Identities = 117/159 (73%), Positives = 136/159 (85%), Gaps = 0/159 (0%)
Query 89 SSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSS 148
S RKQCDF KLLMAGYDLEL N +TQ+FNV FHGP TIYE GVW+VHV LPDDYPF+S
Sbjct 6 SHIRKQCDFTKLLMAGYDLELVNGSTQEFNVTFHGPNGTIYEDGVWQVHVILPDDYPFAS 65
Query 149 PSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSE 208
PSIGF+NK+LHPNVDE+SGSVCLDVIN+TWTP+YSLVNVFDVFLPQLLTYPNP DPLN+E
Sbjct 66 PSIGFMNKMLHPNVDESSGSVCLDVINETWTPIYSLVNVFDVFLPQLLTYPNPSDPLNNE 125
Query 209 AASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQE 247
AA+L+ DK YE+KV+EHV +AS++ WE+ + QE
Sbjct 126 AAALMMLDKTTYEEKVREHVRIHASKEMWEKRRGFTNQE 164
> bbo:BBOV_IV009650 23.m05904; ubiquitin-conjugating enzyme subunit
(EC:6.3.2.19); K10576 ubiquitin-conjugating enzyme E2
H [EC:6.3.2.19]
Length=191
Score = 248 bits (632), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 118/178 (66%), Positives = 140/178 (78%), Gaps = 2/178 (1%)
Query 83 MTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 142
M IS+ RKQ DF KLLMAGYDLEL N N +FNV FHGP TIYE GVW+V VTLP+
Sbjct 1 MNNVSISNVRKQNDFTKLLMAGYDLELINGNMTEFNVTFHGPIGTIYEDGVWKVKVTLPE 60
Query 143 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 202
DYPF+SPSIGFLNK+LHPNVDE SGSVCL+VIN+TWTP+YSLVNVFD FLPQLLTYPNP
Sbjct 61 DYPFASPSIGFLNKMLHPNVDETSGSVCLNVINETWTPIYSLVNVFDTFLPQLLTYPNPS 120
Query 203 DPLNSEAASLLNRDKAQYEQKVKEHVLKYASRDSWEREQAR--KKQEEAQERRREVEE 258
DPLN+EAA LL DK YE+KV+EHV ++AS++ WE+++ K EE +V++
Sbjct 121 DPLNNEAARLLMLDKPTYEKKVREHVKQHASKERWEKQRVNVFKDDEEVDAIISDVDD 178
> ath:AT1G63800 UBC5; UBC5 (ubiquitin-conjugating enzyme 5); ubiquitin-protein
ligase; K10576 ubiquitin-conjugating enzyme
E2 H [EC:6.3.2.19]
Length=185
Score = 204 bits (520), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 99/185 (53%), Positives = 127/185 (68%), Gaps = 11/185 (5%)
Query 90 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSP 149
S R++ D KL+M+ Y +E+ N Q+F V F GPK++IYEGGVW++ V LPD YP+ SP
Sbjct 5 SKRREMDLMKLMMSDYKVEMINDGMQEFFVEFSGPKDSIYEGGVWKIRVELPDAYPYKSP 64
Query 150 SIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEA 209
S+GF+ KI HPNVDE SGSVCLDVINQTW+P++ LVNVF+ FLPQLL YPNP DPLN EA
Sbjct 65 SVGFITKIYHPNVDEMSGSVCLDVINQTWSPMFDLVNVFETFLPQLLLYPNPSDPLNGEA 124
Query 210 ASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEEAQERRREVEEPQLRELSRVDD 269
A+L+ RD+ YEQ+VKE+ KYA K + + +E + E + S DD
Sbjct 125 AALMMRDRPTYEQRVKEYCEKYA-----------KPRADTEEMSSDDEMSEDEYASDGDD 173
Query 270 EADGA 274
E D A
Sbjct 174 EDDVA 178
> ath:AT5G41340 UBC4; UBC4 (UBIQUITIN CONJUGATING ENZYME 4); ubiquitin-protein
ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=187
Score = 199 bits (507), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 88/143 (61%), Positives = 113/143 (79%), Gaps = 0/143 (0%)
Query 90 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSP 149
S R++ D KL+M+ Y +E N Q+F V F+GPK+++Y+GGVW++ V LPD YP+ SP
Sbjct 5 SKRREMDMMKLMMSDYKVETINDGMQEFYVEFNGPKDSLYQGGVWKIRVELPDAYPYKSP 64
Query 150 SIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEA 209
S+GF+ KI HPNVDE SGSVCLDVINQTW+P++ LVNVF+ FLPQLL YPNP DPLN EA
Sbjct 65 SVGFITKIYHPNVDELSGSVCLDVINQTWSPMFDLVNVFETFLPQLLLYPNPSDPLNGEA 124
Query 210 ASLLNRDKAQYEQKVKEHVLKYA 232
A+L+ RD+ YEQ+VKE+ KYA
Sbjct 125 AALMMRDRPAYEQRVKEYCEKYA 147
> ath:AT2G46030 UBC6; UBC6 (ubiquitin-conjugating enzyme 6); ubiquitin-protein
ligase; K10576 ubiquitin-conjugating enzyme
E2 H [EC:6.3.2.19]
Length=183
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 87/143 (60%), Positives = 113/143 (79%), Gaps = 0/143 (0%)
Query 90 SSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSP 149
S R++ D KL+M+ Y ++ N + Q F V FHGP +++Y+GGVW++ V LP+ YP+ SP
Sbjct 5 SKRREMDMMKLMMSDYKVDTVNDDLQMFYVTFHGPTDSLYQGGVWKIKVELPEAYPYKSP 64
Query 150 SIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEA 209
S+GF+NKI HPNVDE+SG+VCLDVINQTW+P++ L+NVF+ FLPQLL YPNP DP N EA
Sbjct 65 SVGFVNKIYHPNVDESSGAVCLDVINQTWSPMFDLINVFESFLPQLLLYPNPSDPFNGEA 124
Query 210 ASLLNRDKAQYEQKVKEHVLKYA 232
ASLL RD+A YE KVKE+ KYA
Sbjct 125 ASLLMRDRAAYELKVKEYCEKYA 147
> sce:YEL012W UBC8, GID3; Ubc8p (EC:6.3.2.19); K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=218
Score = 187 bits (474), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 84/151 (55%), Positives = 116/151 (76%), Gaps = 0/151 (0%)
Query 89 SSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSS 148
S R + D KLLM+ + ++L N + Q+F+V F GPK+T YE GVWR+HV LPD+YP+ S
Sbjct 4 SKRRIETDVMKLLMSDHQVDLINDSMQEFHVKFLGPKDTPYENGVWRLHVELPDNYPYKS 63
Query 149 PSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSE 208
PSIGF+NKI HPN+D ASGS+CLDVIN TW+PLY L+N+ + +P LL PN DPLN+E
Sbjct 64 PSIGFVNKIFHPNIDIASGSICLDVINSTWSPLYDLINIVEWMIPGLLKEPNGSDPLNNE 123
Query 209 AASLLNRDKAQYEQKVKEHVLKYASRDSWER 239
AA+L RDK YE+K+KE++ KYA+++ +++
Sbjct 124 AATLQLRDKKLYEEKIKEYIDKYATKEKYQQ 154
> mmu:100039133 Gm2058; predicted gene 2058; K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=183
Score = 184 bits (468), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 89/167 (53%), Positives = 116/167 (69%), Gaps = 7/167 (4%)
Query 83 MTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 142
M+ R D KL+ + +++ + +F V F+GP+ET YEGGVW+V V LPD
Sbjct 1 MSSPSPGKRRMDTDVVKLIESKHEVTILG-GLNEFVVKFYGPQETPYEGGVWKVRVDLPD 59
Query 143 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 202
YPF SPSIGF+NKI HPN+DEASG+VCLDVINQTWT LY L N+F+ FLPQLL YPNP
Sbjct 60 KYPFKSPSIGFMNKIFHPNIDEASGTVCLDVINQTWTALYDLTNIFESFLPQLLAYPNPI 119
Query 203 DPLNSEAASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEEA 249
DPLN +AA++ +Y+QK+KE++ KYA+ E+A K+QEE
Sbjct 120 DPLNGDAAAMYLHRPEEYKQKIKEYIQKYAT------EEALKEQEEG 160
> cel:Y94H6A.6 ubc-8; UBiquitin Conjugating enzyme family member
(ubc-8); K10576 ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=221
Score = 184 bits (467), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 87/152 (57%), Positives = 107/152 (70%), Gaps = 2/152 (1%)
Query 83 MTQAQ-ISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLP 141
MT A I R CD KL+ +++++ N +F V FHGPK+T YE GVWR+ V +P
Sbjct 1 MTSATAIGKRRIDCDVVKLISHNHEVQIVN-GCSEFIVRFHGPKDTAYENGVWRIRVDMP 59
Query 142 DDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNP 201
D YPF SPSIGFLNKI HPN+DEASG+VCLDVINQ WT LY L N+FD FLPQLLTYPN
Sbjct 60 DKYPFKSPSIGFLNKIFHPNIDEASGTVCLDVINQAWTALYDLTNIFDTFLPQLLTYPNA 119
Query 202 QDPLNSEAASLLNRDKAQYEQKVKEHVLKYAS 233
DPLN EAA L +Y + +E+V+++AS
Sbjct 120 ADPLNGEAARLYIHKPEEYRRTCREYVMRFAS 151
> xla:380288 ube2h, MGC53494; ubiquitin-conjugating enzyme E2H
(UBC8 homolog) (EC:6.3.2.19); K10576 ubiquitin-conjugating
enzyme E2 H [EC:6.3.2.19]
Length=183
Score = 181 bits (460), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 88/166 (53%), Positives = 115/166 (69%), Gaps = 7/166 (4%)
Query 83 MTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 142
M+ R D KL+ + +++ + +F V F+GP+ T YEGGVW+V V LPD
Sbjct 1 MSSPSPGKRRMDTDVVKLIESKHEVTILG-GLNEFIVKFYGPQGTPYEGGVWKVRVDLPD 59
Query 143 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 202
YPF SPSIGF+NKI HPN+DEASG+VCLDVINQTWT LY L N+F+ FLPQLL YPNP
Sbjct 60 KYPFKSPSIGFMNKIFHPNIDEASGTVCLDVINQTWTALYDLTNIFESFLPQLLAYPNPI 119
Query 203 DPLNSEAASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEE 248
DPLN +AA++ +Y+QK+KE++ KYA+ E+A K+QEE
Sbjct 120 DPLNGDAAAMYLHRPEEYKQKIKEYIQKYAT------EEALKEQEE 159
> dre:368425 ube2h, fc07a05, fi24c01, si:bz3c13.2, wu:fc07a05,
wu:fi24c01; ubiquitin-conjugating enzyme E2H (UBC8 homolog,
yeast) (EC:6.3.2.19); K10576 ubiquitin-conjugating enzyme E2
H [EC:6.3.2.19]
Length=183
Score = 181 bits (458), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 88/167 (52%), Positives = 115/167 (68%), Gaps = 7/167 (4%)
Query 83 MTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 142
M+ R D KL+ + +++ + + +F V F+GP+ T YEGGVW+V V LPD
Sbjct 1 MSSPSPGKRRMDTDVVKLIESKHEVTILS-GLNEFVVKFYGPQGTPYEGGVWKVRVDLPD 59
Query 143 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 202
YPF SPSIGF+NKI HPN+DEASG+VCLDVINQTWT LY L N+F+ FLPQLL YPNP
Sbjct 60 KYPFKSPSIGFMNKIFHPNIDEASGTVCLDVINQTWTALYDLTNIFESFLPQLLAYPNPI 119
Query 203 DPLNSEAASLLNRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEEA 249
DPLN +AA++ Y+QK+KE++ KYA+ E+A K+QEE
Sbjct 120 DPLNGDAAAMYLHRPEDYKQKIKEYIQKYAT------EEALKEQEEG 160
> hsa:7328 UBE2H, E2-20K, UBC8, UBCH, UBCH2; ubiquitin-conjugating
enzyme E2H (UBC8 homolog, yeast) (EC:6.3.2.19); K10576
ubiquitin-conjugating enzyme E2 H [EC:6.3.2.19]
Length=113
Score = 122 bits (307), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 57/96 (59%), Positives = 73/96 (76%), Gaps = 6/96 (6%)
Query 154 LNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLL 213
+NKI HPN+DEASG+VCLDVINQTWT LY L N+F+ FLPQLL YPNP DPLN +AA++
Sbjct 1 MNKIFHPNIDEASGTVCLDVINQTWTALYDLTNIFESFLPQLLAYPNPIDPLNGDAAAMY 60
Query 214 NRDKAQYEQKVKEHVLKYASRDSWEREQARKKQEEA 249
+Y+QK+KE++ KYA+ E+A K+QEE
Sbjct 61 LHRPEEYKQKIKEYIQKYAT------EEALKEQEEG 90
> ath:AT1G16890 UBC36; ubiquitin-conjugating enzyme, putative;
K10580 ubiquitin-conjugating enzyme E2 N [EC:6.3.2.19]
Length=153
Score = 96.3 bits (238), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 47/130 (36%), Positives = 77/130 (59%), Gaps = 2/130 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G + +N + FNV+ GP ++ YEGGV+++ + LP++YP ++P + FL KI HPN+D
Sbjct 24 GISASPSEENMRYFNVMILGPTQSPYEGGVFKLELFLPEEYPMAAPKVRFLTKIYHPNID 83
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP DPL+ A ++A+ +
Sbjct 84 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDDPLSENIAKHWKSNEAEAVET 141
Query 224 VKEHVLKYAS 233
KE YAS
Sbjct 142 AKEWTRLYAS 151
> ath:AT5G41700 UBC8; UBC8 (UBIQUITIN CONJUGATING ENZYME 8); protein
binding / ubiquitin-protein ligase
Length=149
Score = 96.3 bits (238), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 67/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP E+ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ + W+P
Sbjct 40 GPAESPYSGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNIN-SNGSICLDILKEQWSPAL 98
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LLT PNP DPL E A + D+A+YE + KYA
Sbjct 99 TISKVL-LSICSLLTDPNPDDPLVPEIAHMYKTDRAKYEATARNWTQKYA 147
> bbo:BBOV_IV007110 23.m05993; ubiquitin-conjugating enzyme E2;
K10580 ubiquitin-conjugating enzyme E2 N [EC:6.3.2.19]
Length=151
Score = 95.9 bits (237), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 49/129 (37%), Positives = 71/129 (55%), Gaps = 2/129 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G ELN N + F + GP T YEGGV+++ + LP+ YP P + FL I HPN+D
Sbjct 20 GIKAELNPGNNRHFYIRMLGPDSTPYEGGVYKLELFLPEQYPMEPPKVRFLTNIYHPNID 79
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ P P DPL++ A DKA E+
Sbjct 80 KL-GRICLDILKDKWSPALQIRTVL-LSVQSLLSAPEPDDPLDTSVADHFKADKADAERL 137
Query 224 VKEHVLKYA 232
+E KYA
Sbjct 138 AREWNEKYA 146
> ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 95.9 bits (237), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 67/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y GGV+ V + P DYPF P + F K+ HPNV+ ++GS+CLD++ + W+P
Sbjct 39 GPSDSPYSGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNVN-SNGSICLDILKEQWSPAL 97
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LLT PNP DPL E A + D+A+YE + KYA
Sbjct 98 TISKVL-LSICSLLTDPNPDDPLVPEIAHMYKTDRAKYESTARSWTQKYA 146
> tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containing
protein (EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=345
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 68/110 (61%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP+++ Y GGV+ +++ P DYPF P + F KI HPN++ + G++CLD++ W+P
Sbjct 237 GPEDSPYSGGVFFLNIHFPSDYPFKPPKVNFTTKIYHPNIN-SQGAICLDILKDQWSPAL 295
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LLT PNP DPL E A L D+ +Y+Q +E KYA
Sbjct 296 TISKVL-LSISSLLTDPNPDDPLVPEIAHLYKSDRMRYDQTAREWSQKYA 344
> sce:YDR059C UBC5; Ubc5p (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 94.7 bits (234), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 47/110 (42%), Positives = 66/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y GGV+ + + P DYPF P + F KI HPN++ +SG++CLD++ W+P
Sbjct 40 GPSDSPYAGGVFFLSIHFPTDYPFKPPKVNFTTKIYHPNIN-SSGNICLDILKDQWSPAL 98
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
+L V + + LLT NP DPL E A + DKA+YE KE KYA
Sbjct 99 TLSKVL-LSICSLLTDANPDDPLVPEIAQIYKTDKAKYEATAKEWTKKYA 147
> ath:AT2G02760 ATUBC2; ATUBC2 (UBIQUITING-CONJUGATING ENZYME
2); ubiquitin-protein ligase; K10573 ubiquitin-conjugating enzyme
E2 A [EC:6.3.2.19]
Length=152
Score = 94.4 bits (233), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/140 (33%), Positives = 79/140 (56%), Gaps = 5/140 (3%)
Query 92 RKQCDFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSS 148
R DF +L AG + N +N V GP +T ++GG +++ + +DYP
Sbjct 8 RLMRDFKRLQQDPPAGISGAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQFSEDYPNKP 67
Query 149 PSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSE 208
P++ F++++ HPN+ A GS+CLD++ W+P+Y + + + LL PNP P NSE
Sbjct 68 PTVRFVSRMFHPNI-YADGSICLDILQNQWSPIYDVAAIL-TSIQSLLCDPNPNSPANSE 125
Query 209 AASLLNRDKAQYEQKVKEHV 228
AA + + K +Y ++V+E V
Sbjct 126 AARMFSESKREYNRRVREVV 145
> ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 94.0 bits (232), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 44/114 (38%), Positives = 67/114 (58%), Gaps = 2/114 (1%)
Query 119 VVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTW 178
GP E+ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ + W
Sbjct 35 ATIMGPPESPYAGGVFLVSIHFPPDYPFKPPKVSFKTKVYHPNIN-SNGSICLDILKEQW 93
Query 179 TPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
+P ++ V + + LLT PNP DPL E A + D+++YE + KYA
Sbjct 94 SPALTISKVL-LSICSLLTDPNPDDPLVPEIAHMYKTDRSKYESTARSWTQKYA 146
> bbo:BBOV_IV010700 23.m05926; ubiquitin-conjugating enzyme; K04649
ubiquitin-conjugating enzyme (huntingtin interacting protein
2) [EC:6.3.2.19]
Length=197
Score = 93.6 bits (231), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 47/158 (29%), Positives = 81/158 (51%), Gaps = 1/158 (0%)
Query 83 MTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPD 142
M + + R+ D K L D N + GP +T YEGG++ +++ +PD
Sbjct 1 MYRQNLRLERELNDIQKELDGPVDAHTVENNIFKWKGYIKGPIQTPYEGGLFILNIDIPD 60
Query 143 DYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQ 202
DYP++ P I F KI HPN+ +G++CLD++ W+P +L + + L++ P P
Sbjct 61 DYPYNPPKIRFETKIWHPNISSETGAICLDILKNEWSPALTLRTAL-LSIQALMSTPEPD 119
Query 203 DPLNSEAASLLNRDKAQYEQKVKEHVLKYASRDSWERE 240
DP ++E A++ R+ ++E+ K +A RE
Sbjct 120 DPQDAEVATMYQRNYDEFERTAKLWTATFAQNRGETRE 157
> ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubiquitin-protein
ligase
Length=178
Score = 93.6 bits (231), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 66/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ + W+P
Sbjct 69 GPSDSPYSGGVFLVTIHFPPDYPFKPPKVAFRTKVFHPNIN-SNGSICLDILKEQWSPAL 127
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LLT PNP DPL E A + DK +YE + KYA
Sbjct 128 TISKVL-LSICSLLTDPNPDDPLVPEIAHMYKTDKNKYESTARTWTQKYA 176
> tpv:TP03_0448 ubiquitin-conjugating enzyme (EC:6.3.2.19); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 93.2 bits (230), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 42/111 (37%), Positives = 69/111 (62%), Gaps = 2/111 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++Y+ GV+ +++ P DYPF P + F K+ HPN++ +G++CLD++ W+P
Sbjct 39 GPHNSLYQNGVYFLNIHFPSDYPFKPPKVAFTTKVYHPNINN-NGAICLDILKDQWSPAL 97
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYAS 233
++ V + + LLT PNP DPL E A + +++ YE V+E V KYA+
Sbjct 98 TISKVL-LSISSLLTDPNPDDPLVPEIAQIYKQNRKLYESTVREWVQKYAT 147
> bbo:BBOV_IV002040 21.m02810; ubiquitin-conjugating enzyme E2
(EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 92.8 bits (229), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 42/111 (37%), Positives = 69/111 (62%), Gaps = 2/111 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++Y+ GV+ +++ P DYPF P + F K+ HPN++ +G++CLD++ W+P
Sbjct 39 GPHNSLYQNGVYFLNIHFPSDYPFKPPKVAFTTKVYHPNINN-NGAICLDILKDQWSPAL 97
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYAS 233
++ V + + LLT PNP DPL E A L +++ YE V++ V KYA+
Sbjct 98 TISKVL-LSISSLLTDPNPDDPLVPEIAQLYKQNRKLYESTVRDWVQKYAT 147
> ath:AT1G14400 UBC1; UBC1 (UBIQUITIN CARRIER PROTEIN 1); ubiquitin-protein
ligase; K10573 ubiquitin-conjugating enzyme E2
A [EC:6.3.2.19]
Length=152
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/136 (33%), Positives = 78/136 (57%), Gaps = 5/136 (3%)
Query 96 DFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIG 152
DF +L AG + N +N V GP +T ++GG +++ + +DYP P++
Sbjct 12 DFKRLQQDPPAGISGAPQDNNIMLWNAVIFGPDDTPWDGGTFKLSLQFSEDYPNKPPTVR 71
Query 153 FLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASL 212
F++++ HPN+ A GS+CLD++ W+P+Y + + + LL PNP P NSEAA +
Sbjct 72 FVSRMFHPNI-YADGSICLDILQNQWSPIYDVAAIL-TSIQSLLCDPNPNSPANSEAARM 129
Query 213 LNRDKAQYEQKVKEHV 228
+ K +Y ++V++ V
Sbjct 130 YSESKREYNRRVRDVV 145
> mmu:93765 Ube2n, 1500026J17Rik, AL022654, BB101821, UBC13; ubiquitin-conjugating
enzyme E2N (EC:6.3.2.19); K10580 ubiquitin-conjugating
enzyme E2 N [EC:6.3.2.19]
Length=152
Score = 92.4 bits (228), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 76/129 (58%), Gaps = 2/129 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G E + N + F+VV GP+++ +EGG +++ + LP++YP ++P + F+ KI HPNVD
Sbjct 22 GIKAEPDESNARYFHVVIAGPQDSPFEGGTFKLELFLPEEYPMAAPKVRFMTKIYHPNVD 81
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP DPL ++ A ++AQ +
Sbjct 82 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDDPLANDVAEQWKTNEAQAIET 139
Query 224 VKEHVLKYA 232
+ YA
Sbjct 140 ARAWTRLYA 148
> hsa:7334 UBE2N, MGC131857, MGC8489, UBC13, UbcH-ben; ubiquitin-conjugating
enzyme E2N (UBC13 homolog, yeast) (EC:6.3.2.19);
K10580 ubiquitin-conjugating enzyme E2 N [EC:6.3.2.19]
Length=152
Score = 92.4 bits (228), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 76/129 (58%), Gaps = 2/129 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G E + N + F+VV GP+++ +EGG +++ + LP++YP ++P + F+ KI HPNVD
Sbjct 22 GIKAEPDESNARYFHVVIAGPQDSPFEGGTFKLELFLPEEYPMAAPKVRFMTKIYHPNVD 81
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP DPL ++ A ++AQ +
Sbjct 82 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDDPLANDVAEQWKTNEAQAIET 139
Query 224 VKEHVLKYA 232
+ YA
Sbjct 140 ARAWTRLYA 148
> ath:AT2G16740 UBC29; UBC29 (ubiquitin-conjugating enzyme 29);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 92.4 bits (228), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 66/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP E+ Y GGV+ V++ P DYPF P + F K+ HPN++ ++G++CLD++ W+P
Sbjct 39 GPNESPYSGGVFLVNIHFPPDYPFKPPKVVFRTKVFHPNIN-SNGNICLDILKDQWSPAL 97
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LLT PNP DPL E A + DK +YE + KYA
Sbjct 98 TISKVL-LSICSLLTDPNPDDPLVPEIAHIYKTDKTKYEAMARSWTQKYA 146
> ath:AT3G08700 UBC12; UBC12 (ubiquitin-conjugating enzyme 12);
small conjugating protein ligase/ ubiquitin-protein ligase;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=149
Score = 92.4 bits (228), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/119 (39%), Positives = 68/119 (57%), Gaps = 8/119 (6%)
Query 120 VFH------GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDV 173
+FH GP ++ Y GGV+ V + DYPF P + F K+ HPN+D + GS+CLD+
Sbjct 31 IFHWQATIMGPHDSPYSGGVFTVSIDFSSDYPFKPPKVNFKTKVYHPNID-SKGSICLDI 89
Query 174 INQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
+ + W+P + V + + LLT PNP DPL E A L DK++YE ++ KYA
Sbjct 90 LKEQWSPAPTTSKVL-LSICSLLTDPNPNDPLVPEIAHLYKVDKSKYESTAQKWTQKYA 147
> dre:393313 ube2nb, MGC63901, ube2nl, zgc:63901; ubiquitin-conjugating
enzyme E2Nb (EC:6.3.2.-); K10580 ubiquitin-conjugating
enzyme E2 N [EC:6.3.2.19]
Length=154
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 77/132 (58%), Gaps = 2/132 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G E + N + F+VV GP+++ +EGG +++ + LP++YP ++P + F+ KI HPNVD
Sbjct 22 GIKAEPDEGNARYFHVVIAGPQDSPFEGGTFKLELFLPEEYPMAAPKVRFMTKIYHPNVD 81
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP DPL ++ A ++AQ +
Sbjct 82 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDDPLANDVAEQWKSNEAQAIET 139
Query 224 VKEHVLKYASRD 235
+ YA +
Sbjct 140 ARTWTRLYAGNN 151
> dre:406807 ube2na, ube2n, wu:fb11b03, wu:fc08b06, zgc:55726;
ubiquitin-conjugating enzyme E2Na (EC:6.3.2.19); K10580 ubiquitin-conjugating
enzyme E2 N [EC:6.3.2.19]
Length=154
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 77/132 (58%), Gaps = 2/132 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G E + N + F+VV GP+++ +EGG +++ + LP++YP ++P + F+ KI HPNVD
Sbjct 22 GIKAEPDEGNARYFHVVIAGPQDSPFEGGTFKLELFLPEEYPMAAPKVRFMTKIYHPNVD 81
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP DPL ++ A ++AQ +
Sbjct 82 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDDPLANDVAEQWKTNEAQAIET 139
Query 224 VKEHVLKYASRD 235
+ YA +
Sbjct 140 ARTWTRLYAGNN 151
> sce:YDR092W UBC13; Ubc13p (EC:6.3.2.19); K10580 ubiquitin-conjugating
enzyme E2 N [EC:6.3.2.19]
Length=153
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 77/146 (52%), Gaps = 12/146 (8%)
Query 102 MAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPN 161
+ G E ++ N + F V GP+++ YE G++ + + LPDDYP +P + FL KI HPN
Sbjct 20 VPGITAEPHDDNLRYFQVTIEGPEQSPYEDGIFELELYLPDDYPMEAPKVRFLTKIYHPN 79
Query 162 VDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYE 221
+D G +CLDV+ W+P + V + + LL PNP DPL ++ A D + E
Sbjct 80 IDRL-GRICLDVLKTNWSPALQIRTVL-LSIQALLASPNPNDPLANDVAE----DWIKNE 133
Query 222 QKVKEHVLKYASRDSWEREQARKKQE 247
Q K A W + A+KK E
Sbjct 134 QGAK------AKAREWTKLYAKKKPE 153
> cel:Y54G2A.31 ubc-13; UBiquitin Conjugating enzyme family member
(ubc-13); K10580 ubiquitin-conjugating enzyme E2 N [EC:6.3.2.19]
Length=151
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 77/129 (59%), Gaps = 2/129 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G + N + F+V+ GP ++ + GGV+++ + LP++YP ++P + F+ KI HPN+D
Sbjct 23 GISANPDESNARYFHVMIAGPDDSPFAGGVFKLELFLPEEYPMAAPKVRFMTKIYHPNID 82
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP+DPL ++ A ++A+ +
Sbjct 83 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPEDPLATDVAEQWKTNEAEAIKT 140
Query 224 VKEHVLKYA 232
K+ + YA
Sbjct 141 AKQWTMNYA 149
> sce:YGL058W RAD6, PSO8, UBC2; Rad6p (EC:6.3.2.19); K10573 ubiquitin-conjugating
enzyme E2 A [EC:6.3.2.19]
Length=172
Score = 91.7 bits (226), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 72/126 (57%), Gaps = 7/126 (5%)
Query 113 NTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLD 172
N +N + GP +T YE G +R+ + ++YP P + FL+++ HPNV A+G +CLD
Sbjct 32 NVMVWNAMIIGPADTPYEDGTFRLLLEFDEEYPNKPPHVKFLSEMFHPNV-YANGEICLD 90
Query 173 VINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ WTP Y + ++ + L PNP P N EAA+L K+QY ++VKE V K
Sbjct 91 ILQNRWTPTYDVASIL-TSIQSLFNDPNPASPANVEAATLFKDHKSQYVKRVKETVEK-- 147
Query 233 SRDSWE 238
SWE
Sbjct 148 ---SWE 150
> mmu:100046119 Mcg1038069; ubiquitin-conjugating enzyme E2 N-like;
K10580 ubiquitin-conjugating enzyme E2 N [EC:6.3.2.19]
Length=203
Score = 91.7 bits (226), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 76/129 (58%), Gaps = 2/129 (1%)
Query 104 GYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVD 163
G E + N + F+VV GP+++ +EGG +++ + LP++YP ++P + F+ KI HPNVD
Sbjct 22 GIKAEPDESNARYFHVVIAGPQDSPFEGGTFKLELFLPEEYPMAAPKVRFMTKIYHPNVD 81
Query 164 EASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQK 223
+ G +CLD++ W+P + V + + LL+ PNP DPL ++ A ++AQ +
Sbjct 82 KL-GRICLDILKDKWSPALQIRTVL-LSIQALLSAPNPDDPLANDVAEQWKTNEAQAIET 139
Query 224 VKEHVLKYA 232
+ YA
Sbjct 140 ARAWTRLYA 148
> cpv:cgd4_210 Ubc1p like ubiquitin-conjugating enzyme E2 fused
to a UBA domain (UBC+UBA) ; K04649 ubiquitin-conjugating enzyme
(huntingtin interacting protein 2) [EC:6.3.2.19]
Length=197
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 44/135 (32%), Positives = 74/135 (54%), Gaps = 2/135 (1%)
Query 109 LNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGS 168
+ N+N +F + GP T YEGG++++ + +P +YP+ P + F+ +I HPN+ +G+
Sbjct 32 IGNRN-DNFLGIIRGPIGTPYEGGIFQLDIIVPKEYPYEPPKVKFITRIWHPNISSQTGA 90
Query 169 VCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHV 228
+CLD++ W+P +L V + + LL+ P P DP ++ ASL D +Y + K
Sbjct 91 ICLDILKDAWSPALTLRTVM-LSIQALLSSPEPNDPQDALVASLYKSDYQEYIETAKSWT 149
Query 229 LKYASRDSWEREQAR 243
YA S E R
Sbjct 150 QMYAKPTSKEERIKR 164
> dre:325470 fc79h10, wu:fc79h10; zgc:55512 (EC:6.3.2.19); K10573
ubiquitin-conjugating enzyme E2 A [EC:6.3.2.19]
Length=152
Score = 91.3 bits (225), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 51/148 (34%), Positives = 83/148 (56%), Gaps = 6/148 (4%)
Query 92 RKQCDFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSS 148
R DF +L AG + N +N V GP+ T +E G +++ V ++YP
Sbjct 8 RLMRDFKRLQEDPPAGVSGAPSENNIMVWNAVIFGPEGTPFEDGTFKLTVEFTEEYPNKP 67
Query 149 PSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSE 208
P++ F++K+ HPNV A GS+CLD++ W+P Y + ++ + LL PNP P NS+
Sbjct 68 PTVRFVSKMFHPNV-YADGSICLDILQNRWSPTYDVSSIL-TSIQSLLDEPNPNSPANSQ 125
Query 209 AASLLNRDKAQYEQKVKEHVLKYASRDS 236
AA L +K +YE++V +++ + RDS
Sbjct 126 AAQLYQENKREYEKRVS-AIVEQSWRDS 152
> sce:YBR082C UBC4; Ubc4p (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 90.9 bits (224), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 65/110 (59%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y GGV+ + + P DYPF P I F KI HPN++ A+G++CLD++ W+P
Sbjct 40 GPADSPYAGGVFFLSIHFPTDYPFKPPKISFTTKIYHPNIN-ANGNICLDILKDQWSPAL 98
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
+L V + + LLT NP DPL E A + D+ +YE +E KYA
Sbjct 99 TLSKVL-LSICSLLTDANPDDPLVPEIAHIYKTDRPKYEATAREWTKKYA 147
> ath:AT5G25760 PEX4; PEX4 (PEROXIN4); protein binding / ubiquitin-protein
ligase; K10689 peroxin-4 [EC:6.3.2.19]
Length=157
Score = 90.9 bits (224), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 75/133 (56%), Gaps = 1/133 (0%)
Query 81 LKMTQAQISSSRKQCDFAKLLMAGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTL 140
++ ++A++ K+ K+ L ++ N + + GP ET YEGGV+++ ++
Sbjct 1 MQASRARLFKEYKEVQREKVADPDIQLICDDTNIFKWTALIKGPSETPYEGGVFQLAFSV 60
Query 141 PDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPN 200
P+ YP P + FL KI HPNV +G +CLD++ W+P ++L +V + L+ +P
Sbjct 61 PEPYPLQPPQVRFLTKIFHPNVHFKTGEICLDILKNAWSPAWTLQSVCRAII-ALMAHPE 119
Query 201 PQDPLNSEAASLL 213
P PLN ++ +LL
Sbjct 120 PDSPLNCDSGNLL 132
> cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 90.9 bits (224), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 43/114 (37%), Positives = 66/114 (57%), Gaps = 2/114 (1%)
Query 119 VVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTW 178
GP E+ Y+GGV+ + + P DYPF P + F +I HPN++ ++GS+CLD++ W
Sbjct 35 ATIMGPPESPYQGGVFFLTIHFPTDYPFKPPKVAFTTRIYHPNIN-SNGSICLDILRSQW 93
Query 179 TPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
+P ++ V + + LL PNP DPL E A + D+ +Y Q +E KYA
Sbjct 94 SPALTISKVL-LSICSLLCDPNPDDPLVPEIARIYKTDRERYNQLAREWTQKYA 146
> mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzyme
E2D 1, UBC4/5 homolog (yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 90.9 bits (224), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 66/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y+GGV+ + V P DYPF P I F KI HPN++ ++GS+CLD++ W+P
Sbjct 39 GPPDSAYQGGVFFLTVHFPTDYPFKPPKIAFTTKIYHPNIN-SNGSICLDILRSQWSPAL 97
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LL PNP DPL + A + DK +Y + +E KYA
Sbjct 98 TVSKVL-LSICSLLCDPNPDDPLVPDIAQIYKSDKEKYNRHAREWTQKYA 146
> hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 90.9 bits (224), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 66/110 (60%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y+GGV+ + V P DYPF P I F KI HPN++ ++GS+CLD++ W+P
Sbjct 39 GPPDSAYQGGVFFLTVHFPTDYPFKPPKIAFTTKIYHPNIN-SNGSICLDILRSQWSPAL 97
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LL PNP DPL + A + DK +Y + +E KYA
Sbjct 98 TVSKVL-LSICSLLCDPNPDDPLVPDIAQIYKSDKEKYNRHAREWTQKYA 146
> cpv:cgd4_570 ubiquitin-conjugating enzyme ; K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=161
Score = 90.5 bits (223), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 68/110 (61%), Gaps = 2/110 (1%)
Query 123 GPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTWTPLY 182
GP ++ Y GGV+ +++ P DYPF P + F KI H N++ ++G++CLD++ + W+P
Sbjct 53 GPDDSSYAGGVFFLNIQFPSDYPFKPPKVNFTTKIYHCNIN-SNGAICLDILKEQWSPAL 111
Query 183 SLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
++ V + + LLT NP DPL E A L D+++YEQ +E KYA
Sbjct 112 TISKVL-LSISSLLTDANPDDPLVPEIAHLYKTDRSKYEQTAREWTQKYA 160
> pfa:PFE1350c ubiquitin conjugating enzyme, putative; K10580
ubiquitin-conjugating enzyme E2 N [EC:6.3.2.19]
Length=152
Score = 90.5 bits (223), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 72/124 (58%), Gaps = 2/124 (1%)
Query 112 KNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCL 171
+N + FN++ +GP T YEGG +++ + LP+ YP P + FL KI HPN+D+ G +CL
Sbjct 29 ENYRHFNILINGPDGTPYEGGTYKLELFLPEQYPMEPPKVRFLTKIYHPNIDKL-GRICL 87
Query 172 DVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKY 231
D++ W+P + V + + LL+ P P DPL+S+ A +DK E ++ Y
Sbjct 88 DILKDKWSPALQIRTVL-LSIQALLSSPEPDDPLDSKVAEHFKQDKNDAEHVARQWNKIY 146
Query 232 ASRD 235
A+ +
Sbjct 147 ANNN 150
> ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 90.5 bits (223), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 42/114 (36%), Positives = 66/114 (57%), Gaps = 2/114 (1%)
Query 119 VVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIGFLNKILHPNVDEASGSVCLDVINQTW 178
GP ++ + GGV+ V + P DYPF P + F K+ HPN++ ++GS+CLD++ + W
Sbjct 35 ATIMGPADSPFAGGVFLVTIHFPPDYPFKPPKVAFRTKVYHPNIN-SNGSICLDILKEQW 93
Query 179 TPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASLLNRDKAQYEQKVKEHVLKYA 232
+P ++ V + + LLT PNP DPL E A + D+ +YE + KYA
Sbjct 94 SPALTVSKVL-LSICSLLTDPNPDDPLVPEIAHIYKTDRVKYESTAQSWTQKYA 146
> dre:797853 ube2a, MGC64109, mp:zf637-2-000771, si:bz46j2.4,
wu:fa01h11; ubiquitin-conjugating enzyme E2A (RAD6 homolog)
(EC:6.3.2.19); K10573 ubiquitin-conjugating enzyme E2 A [EC:6.3.2.19]
Length=152
Score = 90.1 bits (222), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 76/136 (55%), Gaps = 5/136 (3%)
Query 96 DFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIG 152
DF +L AG + N +N V GP+ T +E G +++ + ++YP P++
Sbjct 12 DFKRLQEDPPAGVSGAPSENNIMVWNAVIFGPEGTPFEDGTFKLTIEFTEEYPNKPPTVR 71
Query 153 FLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASL 212
F++K+ HPNV A GS+CLD++ W+P Y + ++ + LL PNP P NS+AA L
Sbjct 72 FVSKMFHPNV-YADGSICLDILQNRWSPTYDVSSIL-TSIQSLLDEPNPNSPANSQAAQL 129
Query 213 LNRDKAQYEQKVKEHV 228
+K +YE++V V
Sbjct 130 YQENKREYEKRVSAIV 145
> xla:398788 ube2a, MGC68540; ubiquitin-conjugating enzyme E2A
(RAD6 homolog) (EC:6.3.2.19); K10573 ubiquitin-conjugating
enzyme E2 A [EC:6.3.2.19]
Length=152
Score = 90.1 bits (222), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 76/136 (55%), Gaps = 5/136 (3%)
Query 96 DFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIG 152
DF +L AG + N +N V GP+ T +E G +++ + ++YP P++
Sbjct 12 DFKRLQEDPPAGVSGAPSENNIMVWNAVIFGPEGTPFEDGTFKLTIEFTEEYPNKPPTVR 71
Query 153 FLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASL 212
F++K+ HPNV A GS+CLD++ W+P Y + ++ + LL PNP P NS+AA L
Sbjct 72 FVSKMFHPNV-YADGSICLDILQNRWSPTYDVSSIL-TSIQSLLDEPNPNSPANSQAAQL 129
Query 213 LNRDKAQYEQKVKEHV 228
+K +YE++V V
Sbjct 130 YQENKREYEKRVSAIV 145
> mmu:22209 Ube2a, HHR6A, HR6A, Mhr6a; ubiquitin-conjugating enzyme
E2A, RAD6 homolog (S. cerevisiae) (EC:6.3.2.19); K10573
ubiquitin-conjugating enzyme E2 A [EC:6.3.2.19]
Length=152
Score = 90.1 bits (222), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 76/136 (55%), Gaps = 5/136 (3%)
Query 96 DFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIG 152
DF +L AG + N +N V GP+ T +E G +++ + ++YP P++
Sbjct 12 DFKRLQEDPPAGVSGAPSENNIMVWNAVIFGPEGTPFEDGTFKLTIEFTEEYPNKPPTVR 71
Query 153 FLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASL 212
F++K+ HPNV A GS+CLD++ W+P Y + ++ + LL PNP P NS+AA L
Sbjct 72 FVSKMFHPNV-YADGSICLDILQNRWSPTYDVSSIL-TSIQSLLDEPNPNSPANSQAAQL 129
Query 213 LNRDKAQYEQKVKEHV 228
+K +YE++V V
Sbjct 130 YQENKREYEKRVSAIV 145
> hsa:7319 UBE2A, HHR6A, RAD6A, UBC2; ubiquitin-conjugating enzyme
E2A (RAD6 homolog) (EC:6.3.2.19); K10573 ubiquitin-conjugating
enzyme E2 A [EC:6.3.2.19]
Length=152
Score = 90.1 bits (222), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 76/136 (55%), Gaps = 5/136 (3%)
Query 96 DFAKLLM---AGYDLELNNKNTQDFNVVFHGPKETIYEGGVWRVHVTLPDDYPFSSPSIG 152
DF +L AG + N +N V GP+ T +E G +++ + ++YP P++
Sbjct 12 DFKRLQEDPPAGVSGAPSENNIMVWNAVIFGPEGTPFEDGTFKLTIEFTEEYPNKPPTVR 71
Query 153 FLNKILHPNVDEASGSVCLDVINQTWTPLYSLVNVFDVFLPQLLTYPNPQDPLNSEAASL 212
F++K+ HPNV A GS+CLD++ W+P Y + ++ + LL PNP P NS+AA L
Sbjct 72 FVSKMFHPNV-YADGSICLDILQNRWSPTYDVSSIL-TSIQSLLDEPNPNSPANSQAAQL 129
Query 213 LNRDKAQYEQKVKEHV 228
+K +YE++V V
Sbjct 130 YQENKREYEKRVSAIV 145
Lambda K H
0.321 0.137 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10720471776
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40