bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1100_orf2
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
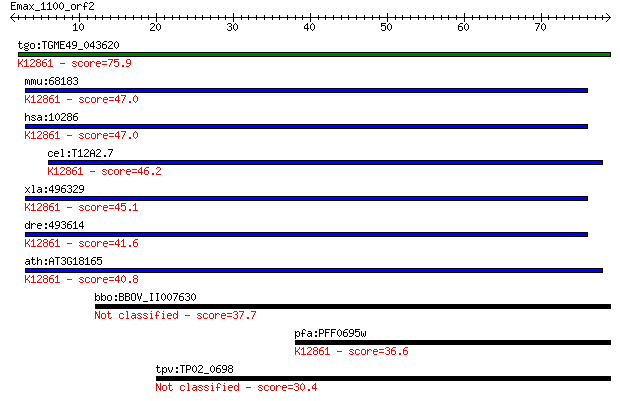
tgo:TGME49_043620 hypothetical protein ; K12861 pre-mRNA-splic... 75.9 3e-14
mmu:68183 Bcas2, 6430539P16Rik, AI132645, C76366, C80030, MGC7... 47.0 1e-05
hsa:10286 BCAS2, DAM1, SPF27, Snt309; breast carcinoma amplifi... 47.0 1e-05
cel:T12A2.7 hypothetical protein; K12861 pre-mRNA-splicing fac... 46.2 2e-05
xla:496329 bcas2; breast carcinoma amplified sequence 2; K1286... 45.1 6e-05
dre:493614 bcas2, cb302, zgc:101730; breast carcinoma amplifie... 41.6 7e-04
ath:AT3G18165 MOS4; MOS4 (Modifier of snc1,4); K12861 pre-mRNA... 40.8 0.001
bbo:BBOV_II007630 18.m06634; hypothetical protein 37.7 0.009
pfa:PFF0695w conserved Plasmodium protein, unknown function; K... 36.6 0.023
tpv:TP02_0698 hypothetical protein 30.4 1.5
> tgo:TGME49_043620 hypothetical protein ; K12861 pre-mRNA-splicing
factor SPF27
Length=358
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 51/77 (66%), Gaps = 1/77 (1%)
Query 2 TMLGKEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHI 61
+++G+E+ R +GEP+ +LDLS+ + P+ K+GD W K++ C+ LL+H +
Sbjct 123 SLVGRELLRLKRGEPMQKLDLSRLYE-AAPAPPKSGDTAEWRKSITTCESLLEHLSLGQT 181
Query 62 NLELMNAHAAASWQRHL 78
NL+LMN HA +SW RHL
Sbjct 182 NLDLMNIHAISSWTRHL 198
> mmu:68183 Bcas2, 6430539P16Rik, AI132645, C76366, C80030, MGC7712;
breast carcinoma amplified sequence 2; K12861 pre-mRNA-splicing
factor SPF27
Length=225
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 3 MLGKEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHIN 62
++ E R A +PI L + +Y PS + D+ AW++ V N L+H A N
Sbjct 65 IMRNEFERLAARQPIELLSMKRY-ELPAPSSGQKNDITAWQECVNNSMAQLEHQAVRIEN 123
Query 63 LELMNAHAAASWQ 75
LELM+ H +W+
Sbjct 124 LELMSQHGCNAWK 136
> hsa:10286 BCAS2, DAM1, SPF27, Snt309; breast carcinoma amplified
sequence 2; K12861 pre-mRNA-splicing factor SPF27
Length=225
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 3 MLGKEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHIN 62
++ E R A +PI L + +Y PS + D+ AW++ V N L+H A N
Sbjct 65 IMRNEFERLAARQPIELLSMKRY-ELPAPSSGQKNDITAWQECVNNSMAQLEHQAVRIEN 123
Query 63 LELMNAHAAASWQ 75
LELM+ H +W+
Sbjct 124 LELMSQHGCNAWK 136
> cel:T12A2.7 hypothetical protein; K12861 pre-mRNA-splicing factor
SPF27
Length=238
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 40/73 (54%), Gaps = 2/73 (2%)
Query 6 KEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHINLEL 65
KE+ R K E + +LD+S+ PS K D K W K + N + +H INLEL
Sbjct 78 KEMDRMKKKEEMGKLDMSR-CELPAPSAVKGVDRKLWAKVLRNAKAQNEHLLMRQINLEL 136
Query 66 MNAHAAASW-QRH 77
M+ +AA S+ QR+
Sbjct 137 MDEYAAESYLQRN 149
> xla:496329 bcas2; breast carcinoma amplified sequence 2; K12861
pre-mRNA-splicing factor SPF27
Length=223
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 3 MLGKEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHIN 62
++ E R + +P+ L + +Y PS + D+ AW++ V N L+H A N
Sbjct 65 IMRNEFERLSARQPLELLSMKRY-ELPAPSSGQRNDITAWQECVNNSMAQLEHQAVRIEN 123
Query 63 LELMNAHAAASWQ 75
LELM+ H +W+
Sbjct 124 LELMSQHGCNAWK 136
> dre:493614 bcas2, cb302, zgc:101730; breast carcinoma amplified
sequence 2; K12861 pre-mRNA-splicing factor SPF27
Length=225
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 3 MLGKEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHIN 62
++ E R A +P+ L + +Y PS + D+ AW+ V N L+H A N
Sbjct 65 IMRNEFERLAARQPMELLSMKRY-ELPAPSSGQKNDMTAWQDCVNNSMAQLEHQAVRIEN 123
Query 63 LELMNAHAAASWQ 75
LELM + +W+
Sbjct 124 LELMAQYGTNAWK 136
> ath:AT3G18165 MOS4; MOS4 (Modifier of snc1,4); K12861 pre-mRNA-splicing
factor SPF27
Length=253
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 0/75 (0%)
Query 3 MLGKEIARKAKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHIN 62
+LGKE R G+P +D +P +K D AW++ + Q+ LQ N
Sbjct 84 VLGKEYERVRAGKPPVRIDFESRYKLEMPPANKRNDDAAWKQYLQKNQRSLQQKLIELEN 143
Query 63 LELMNAHAAASWQRH 77
LELM+ W+++
Sbjct 144 LELMSKLGPELWRQN 158
> bbo:BBOV_II007630 18.m06634; hypothetical protein
Length=195
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 35/67 (52%), Gaps = 8/67 (11%)
Query 12 AKGEPIPELDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHINLELMNAHAA 71
A+GE I LDL KY+ FS D+ + + L ++A A +NLELM+ +
Sbjct 85 ARGEHIQALDLEKYSGFS-----HLDDIDERKGHI---SVLSEYAQGALLNLELMDRYKE 136
Query 72 ASWQRHL 78
+ W RHL
Sbjct 137 SVWLRHL 143
> pfa:PFF0695w conserved Plasmodium protein, unknown function;
K12861 pre-mRNA-splicing factor SPF27
Length=255
Score = 36.6 bits (83), Expect = 0.023, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 38 DVKAWEKTVINCQQLLQHAATAHINLELMNAHAAASWQRHL 78
+++ WEKT+ + +L+++ A IN+ELMN + W H+
Sbjct 138 NIEEWEKTLKKYEIILENSHNALINMELMNKYKEVMWSEHM 178
> tpv:TP02_0698 hypothetical protein
Length=186
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/59 (25%), Positives = 30/59 (50%), Gaps = 8/59 (13%)
Query 20 LDLSKYTSFSVPSGSKAGDVKAWEKTVINCQQLLQHAATAHINLELMNAHAAASWQRHL 78
+D +KY GD K + + + + L++++ + INLELM+ + + W +L
Sbjct 84 MDFTKYDGL--------GDNKDLKAKMSHLKMLMEYSQDSLINLELMDRYKESCWLNYL 134
Lambda K H
0.313 0.125 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40