bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1065_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
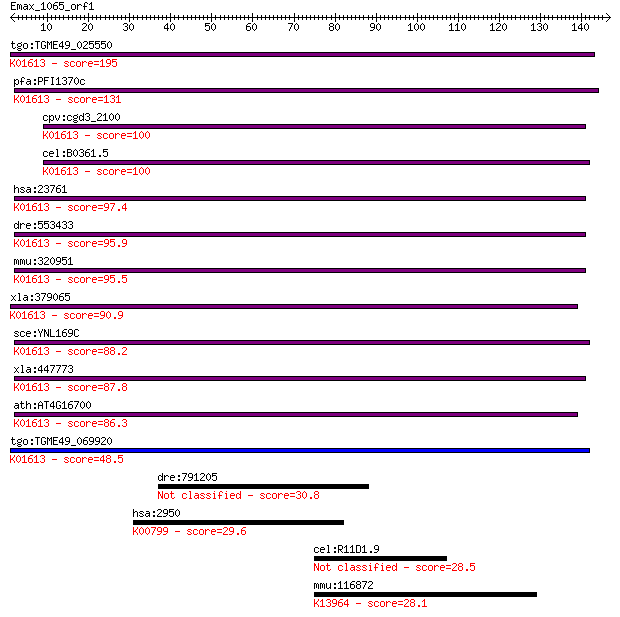
tgo:TGME49_025550 phosphatidylserine decarboxylase proenzyme, ... 195 5e-50
pfa:PFI1370c PfPSD; phosphatidylserine decarboxylase (EC:4.1.1... 131 6e-31
cpv:cgd3_2100 phosphatidylserine decarboxylase ; K01613 phosph... 100 1e-21
cel:B0361.5 psd-1; PhosphatidylSerine Decarboxylase family mem... 100 2e-21
hsa:23761 PISD, DJ858B16, DKFZp566G2246, PSD, PSDC, PSSC, dJ85... 97.4 1e-20
dre:553433 pisd, wu:fd05a08, zgc:158135; phosphatidylserine de... 95.9 4e-20
mmu:320951 Pisd, 9030221M09Rik; phosphatidylserine decarboxyla... 95.5 6e-20
xla:379065 MGC52759; similar to phosphatidylserine decarboxyla... 90.9 1e-18
sce:YNL169C PSD1; Phosphatidylserine decarboxylase of the mito... 88.2 9e-18
xla:447773 pisd, MGC84353; phosphatidylserine decarboxylase (E... 87.8 1e-17
ath:AT4G16700 PSD1; PSD1 (phosphatidylserine decarboxylase 1);... 86.3 3e-17
tgo:TGME49_069920 phosphatidylserine decarboxylase proenzyme, ... 48.5 7e-06
dre:791205 crym, zgc:158843; crystallin, mu 30.8 1.4
hsa:2950 GSTP1, DFN7, FAEES3, GST3, GSTP, PI; glutathione S-tr... 29.6 3.4
cel:R11D1.9 hypothetical protein 28.5 6.8
mmu:116872 Serpinb7, 4631416M05Rik, megsin, ovalbumin; serine ... 28.1 8.9
> tgo:TGME49_025550 phosphatidylserine decarboxylase proenzyme,
putative (EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=337
Score = 195 bits (495), Expect = 5e-50, Method: Compositional matrix adjust.
Identities = 91/142 (64%), Positives = 108/142 (76%), Gaps = 0/142 (0%)
Query 1 CDFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNI 60
F V RHM GE LPVF SFL +FNDIFSV ER+++SG+W G +H+ AVAA NVGNI
Sbjct 181 AKFDVNVLRHMTGETLPVFSSFLKRFNDIFSVNERVVMSGNWKYGCMHMVAVAAYNVGNI 240
Query 61 RLEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGVGSHVGEFRLGSTIVLIFEAPQE 120
R++KEP LRTN+ RVVLRHLGGDV+ RTY +P + VG HVGEFRLGSTIVLIFEAP
Sbjct 241 RIDKEPSLRTNELRVVLRHLGGDVETRTYSRQPFEYSVGQHVGEFRLGSTIVLIFEAPHN 300
Query 121 FEFSVAAGDKIRAGSRLGGVGP 142
F + + G ++R G RLGGVGP
Sbjct 301 FTWDMKPGQEVRVGQRLGGVGP 322
> pfa:PFI1370c PfPSD; phosphatidylserine decarboxylase (EC:4.1.1.65);
K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=353
Score = 131 bits (330), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 59/142 (41%), Positives = 91/142 (64%), Gaps = 1/142 (0%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
+F +RRH+ GE PVF+ N++F + ER+ILSG W GG ++ AA++A NVGNI+
Sbjct 201 NFKYKIRRHISGEVFPVFQGMFKIINNLFDINERVILSGEWKGGHVYYAAISAYNVGNIK 260
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGVGSHVGEFRLGSTIVLIFEAPQEF 121
+ + DL TN R L ++GGD++ + Y D +G VGEF++GS+I++IFE + F
Sbjct 261 IVNDEDLLTNNLRTQLSYMGGDINTKIY-DHYKDLEIGDEVGEFKVGSSIIVIFENKKNF 319
Query 122 EFSVAAGDKIRAGSRLGGVGPP 143
+++V +I G R+GGV P
Sbjct 320 KWNVKPNQQISVGERIGGVDQP 341
> cpv:cgd3_2100 phosphatidylserine decarboxylase ; K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=314
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 56/139 (40%), Positives = 80/139 (57%), Gaps = 7/139 (5%)
Query 9 RHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIRLEKEPDL 68
RH+ GEC PVF+ +K N++FS+ ER+++ W G ++I AVAA V +I+L P+L
Sbjct 170 RHISGECFPVFKGIASKLNNLFSINERVVIKSEWEHGKMYIVAVAAHGVSDIKLFCVPNL 229
Query 69 RTNQDRVVLRHL--GGDVDIRTYLD-----KPLHFGVGSHVGEFRLGSTIVLIFEAPQEF 121
+TNQ L +L G Y D + G +G F LGSTIVLIF+AP+ F
Sbjct 230 KTNQRGSNLNYLRKGKTGQFIEYSDFKNCKNQGKYLKGDELGLFNLGSTIVLIFQAPENF 289
Query 122 EFSVAAGDKIRAGSRLGGV 140
+F V G K++ G +G V
Sbjct 290 KFDVDRGIKLKLGQIIGKV 308
> cel:B0361.5 psd-1; PhosphatidylSerine Decarboxylase family member
(psd-1); K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=377
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 56/133 (42%), Positives = 80/133 (60%), Gaps = 5/133 (3%)
Query 9 RHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIRLEKEPDL 68
RH+ G L V + L+ +F + ER++L+GSW G ++AVAA NVG+I ++ EP L
Sbjct 249 RHVPGLLLSVRPTLLSHVPHLFCLNERVVLNGSWRHGFFSMSAVAATNVGDIVVDAEPSL 308
Query 69 RTNQDRVVLRHLGGDVDIRTYLDKPLHFGVGSHVGEFRLGSTIVLIFEAPQEFEFSVAAG 128
RTN +V R ++ T + P + G VGEFRLGSTIVL+F+AP +F++ AG
Sbjct 309 RTN---IVRRKTQKIMNTETEIHAP--YVSGERVGEFRLGSTIVLVFQAPPTIKFAIKAG 363
Query 129 DKIRAGSRLGGVG 141
D +R G L G
Sbjct 364 DPLRYGQSLVADG 376
> hsa:23761 PISD, DJ858B16, DKFZp566G2246, PSD, PSDC, PSSC, dJ858B16.2;
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613
phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=375
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 78/143 (54%), Gaps = 8/143 (5%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+T + RRH G + V ++F ER++L+G W G + AV A NVG+IR
Sbjct 237 DWTVSHRRHFPGSLMSVNPGMARWIKELFCHNERVVLTGDWKHGFFSLTAVGATNVGSIR 296
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGV----GSHVGEFRLGSTIVLIFEA 117
+ + DL TN R H G + +++ GV G H+GEF LGSTIVLIFEA
Sbjct 297 IYFDRDLHTNSPR----HSKGSYNDFSFVTHTNREGVPMRKGEHLGEFNLGSTIVLIFEA 352
Query 118 PQEFEFSVAAGDKIRAGSRLGGV 140
P++F F + G KIR G LG +
Sbjct 353 PKDFNFQLKTGQKIRFGEALGSL 375
> dre:553433 pisd, wu:fd05a08, zgc:158135; phosphatidylserine
decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=426
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 56/143 (39%), Positives = 79/143 (55%), Gaps = 8/143 (5%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+ RRH G + V ++F ER++LSG W G + AV A NVG+IR
Sbjct 288 DWRVAHRRHFPGALMSVNPGVARWIKELFCHNERVVLSGEWTHGFFSLTAVGATNVGSIR 347
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGV----GSHVGEFRLGSTIVLIFEA 117
+ + +LRTN R + G + +Y+ G+ G H+GEF LGSTIVL+FEA
Sbjct 348 IYFDKELRTNNPR----YNKGTYNDFSYVTNNNQEGISMRKGEHLGEFNLGSTIVLLFEA 403
Query 118 PQEFEFSVAAGDKIRAGSRLGGV 140
P++F F++ AG KIR G LG +
Sbjct 404 PRDFTFNLQAGQKIRFGEALGTM 426
> mmu:320951 Pisd, 9030221M09Rik; phosphatidylserine decarboxylase
(EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=406
Score = 95.5 bits (236), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 56/140 (40%), Positives = 78/140 (55%), Gaps = 2/140 (1%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+T + RRH G + V ++F ER++L+G W G + AV A NVG+IR
Sbjct 268 DWTISHRRHFPGSLMSVNPGMARWIKELFCHNERVVLTGDWKHGFFSLTAVGATNVGSIR 327
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKP-LHFGVGSHVGEFRLGSTIVLIFEAPQE 120
+ + DL TN R + D+ T+ +K + G +GEF LGSTIVLIFEAP++
Sbjct 328 IHFDRDLHTNSPRYS-KGSYNDLSFVTHANKEGIPMRKGEPLGEFNLGSTIVLIFEAPKD 386
Query 121 FEFSVAAGDKIRAGSRLGGV 140
F F + AG KIR G LG +
Sbjct 387 FNFRLKAGQKIRFGEALGSL 406
> xla:379065 MGC52759; similar to phosphatidylserine decarboxylase;
K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=355
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 55/142 (38%), Positives = 72/142 (50%), Gaps = 8/142 (5%)
Query 1 CDFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNI 60
D+ RRH G L V +F ER++LSG W G + AV A NVG+I
Sbjct 216 TDWNVQHRRHFPGALLSVSPHIAHWIPSLFCQNERVVLSGQWQFGFFSLTAVGATNVGSI 275
Query 61 RLEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKP----LHFGVGSHVGEFRLGSTIVLIFE 116
R+ ++ DL TN RH+ G +Y D+ L G +GEF GSTIVLIFE
Sbjct 276 RIYEDQDLHTN----CSRHVKGKYHDYSYTDQYGPEGLTLAKGQPLGEFNFGSTIVLIFE 331
Query 117 APQEFEFSVAAGDKIRAGSRLG 138
P +F+F + AG +I G LG
Sbjct 332 GPLQFKFQIKAGGRIHVGEALG 353
> sce:YNL169C PSD1; Phosphatidylserine decarboxylase of the mitochondrial
inner membrane, converts phosphatidylserine to phosphatidylethanolamine
(EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=500
Score = 88.2 bits (217), Expect = 9e-18, Method: Composition-based stats.
Identities = 52/146 (35%), Positives = 74/146 (50%), Gaps = 8/146 (5%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+ VRRH G+ V F F ++F + ER+ L GSW G + V A NVG+I+
Sbjct 352 DWVCKVRRHFPGDLFSVAPYFQRNFPNLFVLNERVALLGSWKYGFFSMTPVGATNVGSIK 411
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKP------LHFGVGSHVGEFRLGSTIVLIF 115
L + + TN +HL + + + G +G F LGST+VL F
Sbjct 412 LNFDQEFVTNSKSD--KHLEPHTCYQAVYENASKILGGMPLVKGEEMGGFELGSTVVLCF 469
Query 116 EAPQEFEFSVAAGDKIRAGSRLGGVG 141
EAP EF+F V GDK++ G +LG +G
Sbjct 470 EAPTEFKFDVRVGDKVKMGQKLGIIG 495
> xla:447773 pisd, MGC84353; phosphatidylserine decarboxylase
(EC:4.1.1.65); K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=411
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 51/143 (35%), Positives = 76/143 (53%), Gaps = 8/143 (5%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+ RRH G + V ++F ER++L+G W G + AV A NVG+I+
Sbjct 273 DWNVHHRRHFPGSLMSVNPGVAKWIKELFCYNERVVLTGGWKHGFFSLTAVGATNVGSIQ 332
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGV----GSHVGEFRLGSTIVLIFEA 117
+ + DL+TN R + G + +Y+ G+ G +GEF LGSTIVLIFEA
Sbjct 333 IYFDRDLQTNSPR----YSKGSYNDLSYITNNNQDGIVMRKGDQLGEFNLGSTIVLIFEA 388
Query 118 PQEFEFSVAAGDKIRAGSRLGGV 140
P++F F++ G KI G +G +
Sbjct 389 PKDFNFNLKPGQKIHFGEAVGSL 411
> ath:AT4G16700 PSD1; PSD1 (phosphatidylserine decarboxylase 1);
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=453
Score = 86.3 bits (212), Expect = 3e-17, Method: Composition-based stats.
Identities = 54/152 (35%), Positives = 76/152 (50%), Gaps = 18/152 (11%)
Query 2 DFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSWIGGGLHIAAVAACNVGNIR 61
D+ TVRRH G PV +++ ER++L G W G + +AAV A N+G+I
Sbjct 300 DWNATVRRHFAGRLFPVNERATRTIRNLYVENERVVLEGIWKEGFMALAAVGATNIGSIE 359
Query 62 LEKEPDLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGV----GSHVGEFRLGSTIVLIFEA 117
L EP+LRTN+ + L + R Y P G+ G V F +GST+VLIF+A
Sbjct 360 LFIEPELRTNKPKKKLFPTEPPEE-RVY--DPEGLGLRLEKGKEVAVFNMGSTVVLIFQA 416
Query 118 P-----------QEFEFSVAAGDKIRAGSRLG 138
P ++ F V GD++R G LG
Sbjct 417 PTANTPEGSSSSSDYRFCVKQGDRVRVGQALG 448
> tgo:TGME49_069920 phosphatidylserine decarboxylase proenzyme,
putative (EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=711
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 66/153 (43%), Gaps = 21/153 (13%)
Query 1 CDFTQTVRRHMHGECLPVFRSFLAKFNDIFSVQERIILSGSW----IGGGL--HIAAVAA 54
D+T T + ++ G C P + D+ ER L G W G L + VAA
Sbjct 316 ADWTVTSQTYIPG-CTPSVSRRNLEAGDLLHRYERTALIGHWDPEKNGQQLFFSVTMVAA 374
Query 55 CNVGNIRL--EKEP---DLRTNQDRVVLRHLGGDVDIRTYLDKPLHFGVGSHVGEFRLGS 109
VG +RL E+EP +R + VD+ +G FR GS
Sbjct 375 MFVGGLRLSWEEEPLGASMRLGRCTRYTESYEKQVDV--------ELCASQEIGAFRFGS 426
Query 110 TIVLIFEAPQEFEF-SVAAGDKIRAGSRLGGVG 141
T+V+IFEAP++F+ SV + AG G +G
Sbjct 427 TVVMIFEAPEDFDMTSVGQCSHVAAGQPAGYLG 459
> dre:791205 crym, zgc:158843; crystallin, mu
Length=312
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 24/51 (47%), Gaps = 10/51 (19%)
Query 37 ILSGSWIGGGLHIAAVAACNVGNIRLEKEPDLRTNQDRVVLRHLGGDVDIR 87
IL G W+ G HIAAV AC PD R D V++R VD R
Sbjct 209 ILFGEWVKPGAHIAAVGACR---------PDWR-ELDDVLMREAVVYVDSR 249
> hsa:2950 GSTP1, DFN7, FAEES3, GST3, GSTP, PI; glutathione S-transferase
pi 1 (EC:2.5.1.18); K00799 glutathione S-transferase
[EC:2.5.1.18]
Length=210
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 4/51 (7%)
Query 31 SVQERIILSGSWIGGGLHIAAVAACNVGNIRLEKEPDLRTNQDRVVLRHLG 81
S +E ++ +W G L A+C G + ++ DL Q +LRHLG
Sbjct 28 SWKEEVVTVETWQEGSLK----ASCLYGQLPKFQDGDLTLYQSNTILRHLG 74
> cel:R11D1.9 hypothetical protein
Length=187
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 23/37 (62%), Gaps = 5/37 (13%)
Query 75 VVLRHLGGDV-----DIRTYLDKPLHFGVGSHVGEFR 106
V +R++ GD+ D+R+YL++ L + SHV E +
Sbjct 129 VTIRNVDGDIFACENDLRSYLEEHLGHSIASHVDELK 165
> mmu:116872 Serpinb7, 4631416M05Rik, megsin, ovalbumin; serine
(or cysteine) peptidase inhibitor, clade B, member 7; K13964
serpin B7
Length=380
Score = 28.1 bits (61), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 29/63 (46%), Gaps = 9/63 (14%)
Query 75 VVLRHLGGDVDIRTYLDKPLHFGVGSHVG---------EFRLGSTIVLIFEAPQEFEFSV 125
+ L LG D +DK LHF + S G +++L + I + +++E S+
Sbjct 39 LTLIRLGARGDCARQIDKALHFNIPSRQGNSSNNQPGLQYQLKRVLADINSSHKDYELSI 98
Query 126 AAG 128
A G
Sbjct 99 ATG 101
Lambda K H
0.325 0.144 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40