bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
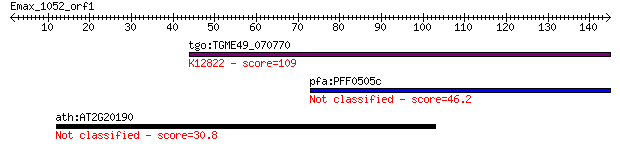
Query= Emax_1052_orf1
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_070770 hypothetical protein ; K12822 RNA-binding pr... 109 3e-24
pfa:PFF0505c conserved Plasmodium protein, unknown function 46.2 3e-05
ath:AT2G20190 CLASP; CLASP (CLIP-ASSOCIATED PROTEIN); binding 30.8 1.6
> tgo:TGME49_070770 hypothetical protein ; K12822 RNA-binding
protein 25
Length=779
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 51/101 (50%), Positives = 71/101 (70%), Gaps = 0/101 (0%)
Query 44 LRKLDPEAIAAAAENSIYRIAPDGTDRRTYIEEDYRPSRRELERLQNAGRREAEYEREFT 103
L+ ++PE AA+ S+YR+ PDG DRR YI EDYRP RRELERL + RR++E+E+EF
Sbjct 375 LKAMNPELAEAASSCSVYRVGPDGVDRRRYITEDYRPCRRELERLHHLERRDSEFEKEFR 434
Query 104 RREAAWKIQEQNLAVDREKELVGEMEISQEDKRRLIEADLR 144
RRE W +E++ ++EKE+ E EI +D++ LI DLR
Sbjct 435 RRELEWITREEDFKQNKEKEVALEQEIRDDDRQHLINQDLR 475
> pfa:PFF0505c conserved Plasmodium protein, unknown function
Length=960
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 41/72 (56%), Gaps = 0/72 (0%)
Query 73 YIEEDYRPSRRELERLQNAGRREAEYEREFTRREAAWKIQEQNLAVDREKELVGEMEISQ 132
YI ++Y+ + RE ERLQ +E + EREF +RE W E+ + D KE + +
Sbjct 416 YISKEYKINWRERERLQKTEYKEKDLEREFYKREKEWIELEEQIKKDTYKEWNKYLLSKK 475
Query 133 EDKRRLIEADLR 144
+D +LIE DL+
Sbjct 476 KDIAKLIEIDLK 487
> ath:AT2G20190 CLASP; CLASP (CLIP-ASSOCIATED PROTEIN); binding
Length=1439
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 45/106 (42%), Gaps = 26/106 (24%)
Query 12 GSSGSNLTNGSS-----SSKTAAAAAADTAAGSAFHRLRKLDPE----AIAAAAENSIYR 62
G G N+++G+ + +T ++A+D L+P+ A+A +NSI R
Sbjct 1092 GGVGQNVSSGTQEKLYQNVRTGISSASDL-----------LNPKDSDYTFASAGQNSISR 1140
Query 63 IAPDGTDRRTYIEEDYRPSRRELERLQNA------GRREAEYEREF 102
+P+G+ I +D P E L GR E E RE
Sbjct 1141 TSPNGSSENIEILDDLSPPHLEKNGLNLTSVDSLEGRHENEVSREL 1186
Lambda K H
0.306 0.121 0.320
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40