bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0990_orf1
Length=325
Score E
Sequences producing significant alignments: (Bits) Value
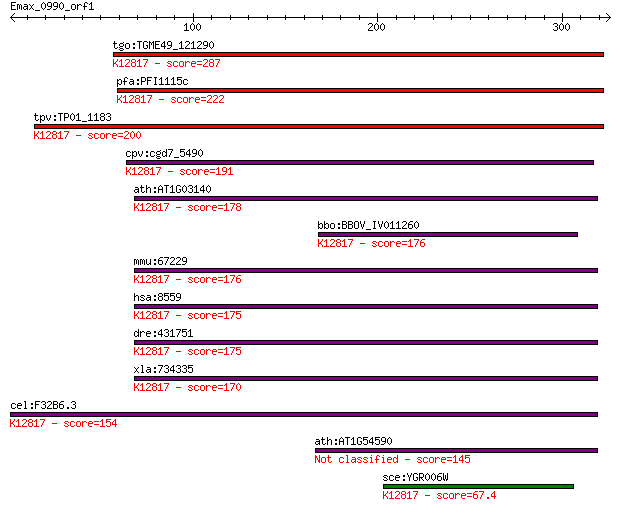
tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K128... 287 3e-77
pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mR... 222 1e-57
tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRN... 200 7e-51
cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-spli... 191 4e-48
ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre... 178 2e-44
bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K128... 176 9e-44
mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing fac... 176 2e-43
hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA proce... 175 2e-43
dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor... 175 2e-43
xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor ... 170 5e-42
cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing fac... 154 4e-37
ath:AT1G54590 splicing factor Prp18 family protein 145 1e-34
sce:YGR006W PRP18; Prp18p; K12817 pre-mRNA-splicing factor 18 67.4
> tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K12817
pre-mRNA-splicing factor 18
Length=373
Score = 287 bits (735), Expect = 3e-77, Method: Compositional matrix adjust.
Identities = 158/331 (47%), Positives = 198/331 (59%), Gaps = 66/331 (19%)
Query 57 TDEEREPPVELQEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFH 116
DEE EPP+ QEIF++LRK+K+P+TLF E+ W+RY RLC+LE+ +DE+ GQ+N F
Sbjct 39 NDEETEPPLPKQEIFKKLRKMKEPVTLFGESDWQRYTRLCELELLHHEDELMGGQRNAFL 98
Query 117 AMQREGE--------------------EEEEFEEESKPVPSSEKGEAEKSDA-------- 148
Q + E E+ + +E+S P + G E DA
Sbjct 99 NPQDDDEEDEDNLKERASSPRRRPPRREQGDADEDSGSSPHARPGGEEAPDAGSRKQGYL 158
Query 149 ---------------AAATQNAKSKETT-----------------------EQTKEAAVI 170
AAA + K+ T + +KE V
Sbjct 159 SSSSERDSGAPADKEAAAEKRTKATNTAGSGSDEEDGTDGSGVGRGGQQERQPSKETVVT 218
Query 171 GWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEK 230
W R ML WE ELK+R E++K +AEG T+L RQT+KDLKPL +KL+H++LE DIL+K
Sbjct 219 RWIRTMLQEWEAELKARPEEKKNSAEGKLMTSLQRQTRKDLKPLLRKLRHKELENDILQK 278
Query 231 LFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDE 290
L +V CEERKYR AH +MLLAIGNAAWP+GVTMVGIHER GRSKL +SQVAHILNDE
Sbjct 279 LHIMVQCCEERKYRSAHDTYMLLAIGNAAWPVGVTMVGIHERVGRSKLFSSQVAHILNDE 338
Query 291 TTRKYIQMFKRLMSFAQRKFPASPSQTILAS 321
TTRKYIQMFKRLMSF QR++PA PSQTI S
Sbjct 339 TTRKYIQMFKRLMSFCQRRYPADPSQTISLS 369
> pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mRNA-splicing
factor 18
Length=343
Score = 222 bits (566), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 125/272 (45%), Positives = 176/272 (64%), Gaps = 17/272 (6%)
Query 59 EEREPPVEL----QEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNV 114
E E +E+ ++I LR+LK+PI LF ET +RY RL +L++ +E+ ++N+
Sbjct 76 ENEETNIEITLSNKQIIMLLRQLKEPIRLFGETDLQRYNRLKELKMN--KNELKINEQNI 133
Query 115 FHAMQREGEEEEEFEE-ESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQ----TKEAAV 169
F + R +E+ + E EKG ++K D + + KE E+ KE +
Sbjct 134 FGDVLRGRLKEDSLDLIEDNIEDEIEKG-SDKKDKNNSNVSISEKENNEKGKKIDKEKII 192
Query 170 IGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILE 229
W ++ + W EE+++ + +K QAT L QT KDLKPL KKLK + LE+DIL+
Sbjct 193 HEWIKRTMKEWNEEIENIVDSKKKIK---QATYL--QTHKDLKPLEKKLKQKTLESDILD 247
Query 230 KLFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILND 289
K+++IVS C+E+ ++ AH A+MLLAIGNAAWPMGVTMVGIHERAGRSK+ S+VAHILND
Sbjct 248 KIYNIVSCCQEKNFKAAHDAYMLLAIGNAAWPMGVTMVGIHERAGRSKIYASEVAHILND 307
Query 290 ETTRKYIQMFKRLMSFAQRKFPASPSQTILAS 321
ETTRKYIQM KRL+SF QRK+ +PS+ + S
Sbjct 308 ETTRKYIQMIKRLLSFCQRKYCTNPSEAVNLS 339
> tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRNA-splicing
factor 18
Length=327
Score = 200 bits (508), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 125/315 (39%), Positives = 173/315 (54%), Gaps = 31/315 (9%)
Query 14 KRKAELEQLAELQDRIKARPFQKNPEAEAAAAATTECADTTEGTDEEREPP--VELQEIF 71
KRK E ++ E Q+++K + N E +T D +P V ++E+
Sbjct 33 KRKLEAQEQLEKQNQLKKKLINDN------FKKIEEFYETNTNLDSTHDPTQDVSVEEVV 86
Query 72 RRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVF--HAMQREGEEEEEFE 129
+RLRKL+QPI F E+ +R RL E IDD E +N++ M R ++
Sbjct 87 KRLRKLRQPIVFFGESHKERCRRLFSQETD-IDD--LEANQNIYVDAIMGRNNYLISKYT 143
Query 130 EESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEAAVIGWARKMLSLWEEELKSRSE 189
+++ + + D + Q + +K V W +ML WE +
Sbjct 144 HKNQ-------SQLDFFDDFSGLQ--------QDSKHYKVFEWVSRMLREWESRIVESKG 188
Query 190 D---EKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVSLCEERKYRDA 246
D E E + A+ QTKKD+KPL K +K L+ +IL+K+ IV+ C +++ A
Sbjct 189 DLIKEGKEFEAKKNEAMLVQTKKDIKPLLKLIKSNKLDQEILDKMEQIVNHCNNGQFKKA 248
Query 247 HGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQMFKRLMSFA 306
H +MLLAIGNAAWPMGVTMVGIHERAGRSK+ TS+VAHILNDETTRKYIQMFKRL+SF+
Sbjct 249 HDIYMLLAIGNAAWPMGVTMVGIHERAGRSKIFTSEVAHILNDETTRKYIQMFKRLISFS 308
Query 307 QRKFPASPSQTILAS 321
Q K+ PSQ I S
Sbjct 309 QSKYAKDPSQIIQIS 323
> cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-splicing
factor 18
Length=337
Score = 191 bits (484), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 107/257 (41%), Positives = 153/257 (59%), Gaps = 21/257 (8%)
Query 64 PVELQEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQV-IDDEMTEGQKNVF-HAMQRE 121
P++ +E +RLR L +PITLF E +RY RL +LE + ++EM GQ+N+F H
Sbjct 94 PIQNEETIKRLRLLGEPITLFGEDDNERYNRLRRLEFKGRTNEEMNIGQQNIFLHGF--- 150
Query 122 GEEEEEFEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEAAVIGWARKMLSLWE 181
+ F S ++ E+ + N K ++ W L WE
Sbjct 151 ---DPNFSLYSVSDDHTQVEIYEEFEHIEGESNQK-----------FILRWIDTQLKEWE 196
Query 182 EELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEAD--ILEKLFDIVSLCE 239
+ LK R + + T +GMQ +A + QTK+D++PL L+ D + D +L+KLF+IV+LC
Sbjct 197 KLLKCRKKADSETEKGMQDSAQYYQTKRDIQPLVNSLEASDGKVDKEVLDKLFEIVTLCN 256
Query 240 ERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQMF 299
+R Y A ++ LAIGNA WPMGVTMVGIHERAGR+K+ +S +AH+LNDETTRKYIQMF
Sbjct 257 QRDYNKAQDKYIELAIGNAPWPMGVTMVGIHERAGRTKIFSSHIAHVLNDETTRKYIQMF 316
Query 300 KRLMSFAQRKFPASPSQ 316
KRL++ + K P+ Q
Sbjct 317 KRLVTHCESKRPSREQQ 333
> ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre-mRNA-splicing
factor 18
Length=420
Score = 178 bits (452), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 108/272 (39%), Positives = 148/272 (54%), Gaps = 21/272 (7%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRL---CKLEVQVIDDEMTEGQKNVF-------HA 117
QE+ RRLR LKQP+TLF E R RL K + +D +MTEGQ N F
Sbjct 107 QEVIRRLRFLKQPMTLFGEDDQSRLDRLKYVLKEGLFEVDSDMTEGQTNDFLRDIAELKK 166
Query 118 MQREG----------EEEEEFEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQ-TKE 166
Q+ G +E E + E E SD A + K E E
Sbjct 167 RQKSGMMGDRKRKSRDERGRDEGDRGETREDELSGGESSDVDADKDMKRLKANFEDLCDE 226
Query 167 AAVIGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEAD 226
++ + +K+L W++EL + E+ TA+G Q A +Q + L PLF + + L AD
Sbjct 227 DKILVFYKKLLIEWKQELDAMENTERRTAKGKQMVATFKQCARYLVPLFNLCRKKGLPAD 286
Query 227 ILEKLFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHI 286
I + L +V+ C +R Y A ++ LAIGNA WP+GVTMVGIHER+ R K+ T+ VAHI
Sbjct 287 IRQALMVMVNHCIKRDYLAAMDHYIKLAIGNAPWPIGVTMVGIHERSAREKIYTNSVAHI 346
Query 287 LNDETTRKYIQMFKRLMSFAQRKFPASPSQTI 318
+NDETTRKY+Q KRLM+F QR++P PS+ +
Sbjct 347 MNDETTRKYLQSVKRLMTFCQRRYPTMPSKAV 378
> bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K12817
pre-mRNA-splicing factor 18
Length=285
Score = 176 bits (447), Expect = 9e-44, Method: Compositional matrix adjust.
Identities = 87/147 (59%), Positives = 109/147 (74%), Gaps = 7/147 (4%)
Query 168 AVIG----WARKMLSLWEEELKSRSED---EKATAEGMQATALHRQTKKDLKPLFKKLKH 220
A+IG W L+ WE+ + S+ E+ E AE + A+ QTKKDL+PL LK
Sbjct 137 AIIGRIASWVSTTLNKWEQHILSQREEFLQEGNVAEARRRDAMLAQTKKDLQPLINMLKS 196
Query 221 RDLEADILEKLFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT 280
+ LE +ILEK++++V CE++ Y+ AH A+MLLAIGNAAWPMGVTMVGIHERAGRSK+ T
Sbjct 197 QTLEEEILEKMYNVVICCEKKDYQAAHEAYMLLAIGNAAWPMGVTMVGIHERAGRSKIFT 256
Query 281 SQVAHILNDETTRKYIQMFKRLMSFAQ 307
S+VAHILNDE TRKYIQM KRL+SFAQ
Sbjct 257 SEVAHILNDEKTRKYIQMIKRLLSFAQ 283
> mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 176 bits (445), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 97/262 (37%), Positives = 150/262 (57%), Gaps = 17/262 (6%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFHAMQREGEEEEE 127
QE+ RRLR+ +PI LF ET + + RL K+E+ + E+ +G +N A + +++
Sbjct 82 QEVIRRLRERGEPIRLFGETDYDAFQRLRKIEI--LTPEVNKGLRNDLKAALDKIDQQYL 139
Query 128 FEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEA-----------AVIGWARKM 176
E V E GE + + + + E E E+ + + + +
Sbjct 140 NE----IVGGQEPGEEDTQNDLKVHEENTTIEELEALGESLGKGDDHKDMDIITKFLKFL 195
Query 177 LSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVS 236
L +W +EL +R + K + +G +A +QT+ L+PLF+KL+ R+L ADI E + DI+
Sbjct 196 LGVWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESITDIIK 255
Query 237 LCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYI 296
+R+Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET RKYI
Sbjct 256 FMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYI 315
Query 297 QMFKRLMSFAQRKFPASPSQTI 318
Q KRLM+ Q+ FP PS+ +
Sbjct 316 QGLKRLMTICQKHFPTDPSKCV 337
> hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA processing
factor 18 homolog (S. cerevisiae); K12817 pre-mRNA-splicing
factor 18
Length=342
Score = 175 bits (444), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 97/262 (37%), Positives = 150/262 (57%), Gaps = 17/262 (6%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFHAMQREGEEEEE 127
QE+ RRLR+ +PI LF ET + + RL K+E+ + E+ +G +N A + +++
Sbjct 82 QEVIRRLRERGEPIRLFGETDYDAFQRLRKIEI--LTPEVNKGLRNDLKAALDKIDQQYL 139
Query 128 FEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEA-----------AVIGWARKM 176
E V E GE + + + + E E E+ + + + +
Sbjct 140 NE----IVGGQEPGEEDTQNDLKVHEENTTIEELEALGESLGKGDDHKDMDIITKFLKFL 195
Query 177 LSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVS 236
L +W +EL +R + K + +G +A +QT+ L+PLF+KL+ R+L ADI E + DI+
Sbjct 196 LGVWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESITDIIK 255
Query 237 LCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYI 296
+R+Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET RKYI
Sbjct 256 FMLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYI 315
Query 297 QMFKRLMSFAQRKFPASPSQTI 318
Q KRLM+ Q+ FP PS+ +
Sbjct 316 QGLKRLMTICQKHFPTDPSKCV 337
> dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 175 bits (443), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 99/263 (37%), Positives = 149/263 (56%), Gaps = 19/263 (7%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFHAMQRE------ 121
QE+ RRLR+ +PI LF E+ + + RL K+E+ + E+ +G +N A +
Sbjct 82 QEVIRRLRERGEPIRLFGESDYDAFQRLRKIEI--LAPEVNKGLRNDLKAAMDKIDQQYL 139
Query 122 -----GEEEEEFEEESKPVPSSEKGEAEKSDAAAATQNAKSKETTEQTKEAAVIGWA-RK 175
G E E + + E E+ +A A S ++ +I R
Sbjct 140 NEIVGGAEGAELDTQHDLKVHEENTTIEELEALGA-----SLGNGNDVRDMDIINKVLRF 194
Query 176 MLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIV 235
+L +W ++L SR + K + +G A+A +QT+ L+PLF+KL+ ++L ADI E + DI+
Sbjct 195 LLGVWAKDLNSREDHIKRSVQGKLASATQKQTESYLEPLFRKLRKKNLPADIKESITDII 254
Query 236 SLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKY 295
ER+Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET RKY
Sbjct 255 KFMLEREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKY 314
Query 296 IQMFKRLMSFAQRKFPASPSQTI 318
IQ KRLM+ Q+ FP PS+ +
Sbjct 315 IQGLKRLMTICQKHFPTDPSKCV 337
> xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor
18 homolog; K12817 pre-mRNA-splicing factor 18
Length=342
Score = 170 bits (431), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 98/261 (37%), Positives = 152/261 (58%), Gaps = 15/261 (5%)
Query 68 QEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFHAMQREGEEEEE 127
QE+ RRLR+ +PI LF ET ++ + RL K+E+ + E+ +G +N A + +++
Sbjct 82 QEVIRRLRERGEPIRLFGETDYETFQRLRKIEI--LAPEVNKGLRNDLKAAL-DKIDQQY 138
Query 128 FEEESKPVPSSEKGEAEKSDAAAATQNAKSKET---------TEQTKEAAVIGWARK-ML 177
F E +++ E ++D +N +E + K+ I K +L
Sbjct 139 FNELVAGQETTD--EDTQNDLKVHEENTTIEELEVLGECLGQGDDNKDMDTINKVLKFLL 196
Query 178 SLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEADILEKLFDIVSL 237
+W +EL +R + K + G A+A +QT+ LKPLF+KL+ ++L ADI E + DI+
Sbjct 197 GVWAKELNAREDYVKRSVHGKLASATQKQTESYLKPLFRKLRKKNLPADIKESITDIIKF 256
Query 238 CEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAHILNDETTRKYIQ 297
+R+Y A+ A++ +AIGNA WP+GVTMVGIH R GR K+ + VAH+LNDET RKYIQ
Sbjct 257 MLQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQ 316
Query 298 MFKRLMSFAQRKFPASPSQTI 318
KRLM+ Q+ F PS+ +
Sbjct 317 GLKRLMTICQKYFSTDPSKCV 337
> cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing factor
18
Length=352
Score = 154 bits (390), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 113/333 (33%), Positives = 161/333 (48%), Gaps = 44/333 (13%)
Query 1 QEKLKEKEELADRKRKAELEQLAELQDRIKARPFQKNPEAEAAAAATTECADTTEGTDEE 60
QE ++++E+A +KRK + E L E R K P + E + E
Sbjct 42 QEYERKQQEIASKKRKVDDEILQESSSRTKPAPVENESEID------------------E 83
Query 61 REPPVELQEIFRRLRKLKQPITLFAETPWKRYCRLCKLEVQVIDDEMTEGQKNVFH-AMQ 119
+ P + EI RLR+ PI LF ET RL +LE+ D + EG +N AM+
Sbjct 84 KTP---MSEIQTRLRQRNHPIMLFGETDIDVRKRLHQLELAQPD--LNEGWENELQTAMK 138
Query 120 REGEEEEEFEEESKPVPSSEKGEAEKSDAAAAT-------QNAKSKETTEQTKEA----- 167
G+E + K V A + D A +N + T +
Sbjct 139 VIGKEMD------KAVVEGTADSATRHDIALPQGYEEDNWKNIEHNSTLLSVDDDLKRDC 192
Query 168 -AVIGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEAD 226
++ R +L+ W ++L R D K TA+GM A H+QT LK L ++ + D
Sbjct 193 DIILSICRYILARWAKDLNDRPLDVKKTAQGMHEAAHHKQTMMHLKSLMTSMERYNCNND 252
Query 227 ILEKLFDIVSL-CEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNTSQVAH 285
I L I L +R Y +A+ A+M +AIGNA WP+GVT GIH+R G +K S +AH
Sbjct 253 IRHHLAKICRLLVIDRNYLEANNAYMEMAIGNAPWPVGVTRSGIHQRPGSAKSYVSNIAH 312
Query 286 ILNDETTRKYIQMFKRLMSFAQRKFPASPSQTI 318
+LNDET RKYIQ FKRLM+ Q FP PS+++
Sbjct 313 VLNDETQRKYIQAFKRLMTKMQEYFPTDPSKSV 345
> ath:AT1G54590 splicing factor Prp18 family protein
Length=256
Score = 145 bits (367), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 73/154 (47%), Positives = 104/154 (67%), Gaps = 1/154 (0%)
Query 166 EAAVIGWARKMLSLWEEELKSRSEDEKATAEGMQATALHRQTKKDLKPLFKKLKHRDLEA 225
E ++ + +K+L W++EL++ E+ TA G Q A Q + L PLF +++ L A
Sbjct 61 EDKILVFCKKLLLEWKQELEAMENTERRTAIGKQMLATFNQCARYLTPLFHLCRNKCLPA 120
Query 226 DILEKLFDIVSLCEERKYRDAHGAFMLLAIGNAAWPMGVTMVGIHERAGRSKLNT-SQVA 284
DI + L +V+ +R Y DA F+ LAIGNA WP+GVTMVGIHER+ R K++T S VA
Sbjct 121 DIRQGLMVMVNCWIKRDYLDATAQFIKLAIGNAPWPIGVTMVGIHERSAREKISTSSSVA 180
Query 285 HILNDETTRKYIQMFKRLMSFAQRKFPASPSQTI 318
HI+N+ETTRKY+Q KRLM+F QR++ A PS++I
Sbjct 181 HIMNNETTRKYLQSVKRLMTFCQRRYSALPSKSI 214
> sce:YGR006W PRP18; Prp18p; K12817 pre-mRNA-splicing factor 18
Length=251
Score = 67.4 bits (163), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 62/105 (59%), Gaps = 2/105 (1%)
Query 203 LHRQTKKDLKPLFKKLKHRDLEADILEKLFDIV-SLCEERKYRDAHGAFMLLAIGNAAWP 261
L TKK L PL +L+ L D+L L ++ L + ++ A ++M L+IGN AWP
Sbjct 135 LFLDTKKALFPLLLQLRRNQLAPDLLISLATVLYHLQQPKEINLAVQSYMKLSIGNVAWP 194
Query 262 MGVTMVGIHERAGRSKLNTS-QVAHILNDETTRKYIQMFKRLMSF 305
+GVT VGIH R+ SK+ A+I+ DE TR +I KRL++F
Sbjct 195 IGVTSVGIHARSAHSKIQGGRNAANIMIDERTRLWITSIKRLITF 239
Lambda K H
0.312 0.126 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13458468080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40