bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0988_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
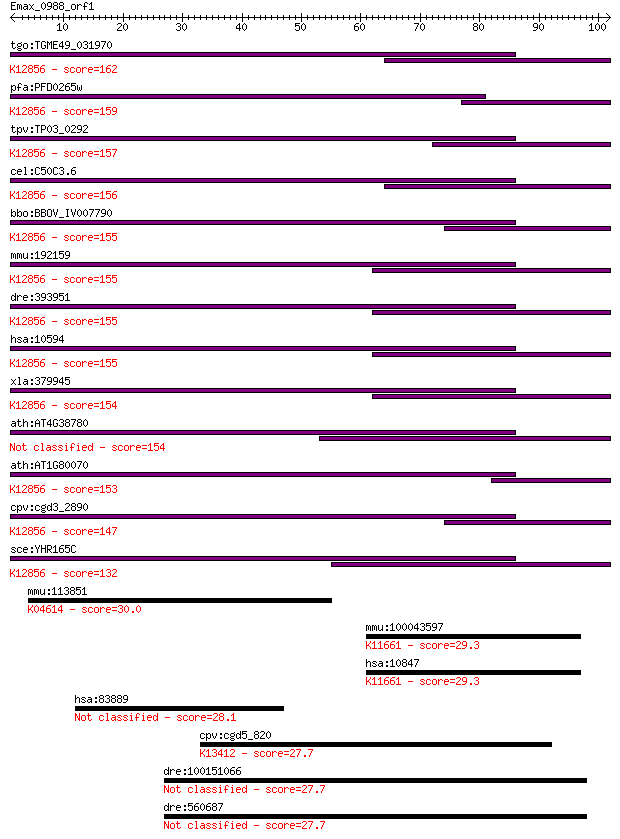
tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ; K1... 162 3e-40
pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mR... 159 3e-39
tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing... 157 9e-39
cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family ... 156 1e-38
bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12... 155 3e-38
mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfpr... 155 4e-38
dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2... 155 4e-38
hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA proce... 155 4e-38
xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13; P... 154 5e-38
ath:AT4G38780 splicing factor, putative 154 1e-37
ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mR... 153 1e-37
cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processin... 147 8e-36
sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component of... 132 3e-31
mmu:113851 Vmn1r54, V1ra9, VN7; vomeronasal 1 receptor 54; K04... 30.0 1.8
mmu:100043597 Srcap, B930091H02Rik, D030022P06Rik, F630004O05R... 29.3 3.0
hsa:10847 SRCAP, DOMO1, EAF1, FLJ44499, KIAA0309, SWR1; Snf2-r... 29.3 3.0
hsa:83889 WDR87, FLJ52608, NYD-SP11; WD repeat domain 87 28.1
cpv:cgd5_820 calcium/calmodulin dependent protein kinase with ... 27.7 8.5
dre:100151066 MGC195210; cardiomyopathy associated 5 like 27.7 8.6
dre:560687 cmya5, cmya5l; cardiomyopathy associated 5 27.7
> tgo:TGME49_031970 pre-mRNA splicing factor PRP8, putative ;
K12856 pre-mRNA-processing factor 8
Length=2538
Score = 162 bits (410), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 77/87 (88%), Positives = 82/87 (94%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FRVFK+TKFFQ TE+DWVE GLQV+RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 675 FRVFKSTKFFQCTELDWVEVGLQVARQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 734
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 735 KERKKSRFGNAFHLCREILRLTKLVVD 761
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 30/43 (69%), Gaps = 5/43 (11%)
Query 64 KKSRFGNA-----FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
+KS+ G + E LRKLIKDHGDM+S+K+RHD+RVY
Sbjct 246 EKSKLGTGQDTTKEEMPPEHLRKLIKDHGDMTSKKFRHDKRVY 288
> pfa:PFD0265w pre-mRNA splicing factor, putative; K12856 pre-mRNA-processing
factor 8
Length=3136
Score = 159 bits (401), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 73/80 (91%), Positives = 77/80 (96%), Gaps = 0/80 (0%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR+FK+TKFFQ TE+DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 1018 FRIFKSTKFFQCTEMDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 1077
Query 61 KERKKSRFGNAFHLCREILR 80
KERKKSRFGNAFHLCREILR
Sbjct 1078 KERKKSRFGNAFHLCREILR 1097
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/25 (68%), Positives = 24/25 (96%), Gaps = 0/25 (0%)
Query 77 EILRKLIKDHGDMSSRKYRHDRRVY 101
E LRK++K+HGDMS++KYR+D+RVY
Sbjct 410 EHLRKIVKEHGDMSNKKYRYDKRVY 434
> tpv:TP03_0292 splicing factor Prp8; K12856 pre-mRNA-processing
factor 8
Length=2736
Score = 157 bits (396), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 75/87 (86%), Positives = 80/87 (91%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
F+VF+ TKFFQ TE+DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 887 FKVFRGTKFFQSTELDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 946
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 947 KERKKSRFGNAFHLCREILRLTKLVVD 973
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 18/30 (60%), Positives = 25/30 (83%), Gaps = 0/30 (0%)
Query 72 FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
+ E +RK+I+DHGDMSSR+YR+D+RVY
Sbjct 278 LQMPPEHVRKVIRDHGDMSSRRYRYDKRVY 307
> cel:C50C3.6 prp-8; yeast PRP (splicing factor) related family
member (prp-8); K12856 pre-mRNA-processing factor 8
Length=2329
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 76/87 (87%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T +DWVEAGLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 465 FRSFKATKFFQTTTLDWVEAGLQVLRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 524
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 525 KERKKSRFGNAFHLCREILRLTKLVVD 551
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 31/42 (73%), Gaps = 4/42 (9%)
Query 64 KKSRFGNA----FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
+K +FG + + E +RK+I+DHGDM+SRKYRHD+RVY
Sbjct 37 EKKKFGMSDTQKEEMPPEHVRKVIRDHGDMTSRKYRHDKRVY 78
> bbo:BBOV_IV007790 23.m06497; processing splicing factor 8; K12856
pre-mRNA-processing factor 8
Length=2343
Score = 155 bits (392), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
F+VF+ TKFFQ TE+DWVEAGLQV QGYNMLNLLIHRKNL YLHLDYNFNL+PVKTLTT
Sbjct 496 FKVFRATKFFQTTELDWVEAGLQVCHQGYNMLNLLIHRKNLTYLHLDYNFNLKPVKTLTT 555
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 556 KERKKSRFGNAFHLCREILRLTKLVVD 582
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 17/28 (60%), Positives = 24/28 (85%), Gaps = 0/28 (0%)
Query 74 LCREILRKLIKDHGDMSSRKYRHDRRVY 101
+ E LRK+ ++HGDMS+RKYR+D+RVY
Sbjct 56 MPPEHLRKVFREHGDMSNRKYRYDKRVY 83
> mmu:192159 Prpf8, AU019467, D11Bwg0410e, DBF3/PRP8, Prp8, Sfprp8l;
pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T++DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 473 FRSFKATKFFQSTKLDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 532
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCRE+LR KL+ D
Sbjct 533 KERKKSRFGNAFHLCREVLRLTKLVVD 559
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 32/42 (76%), Gaps = 2/42 (4%)
Query 62 ERKKSRFGNA--FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
E++K F +A + E +RK+I+DHGDM++RK+RHD+RVY
Sbjct 47 EKRKFGFVDAQKEDMPPEHVRKIIRDHGDMTNRKFRHDKRVY 88
> dre:393951 prpf8, MGC162871, MGC56504, id:ibd1257, ik:tdsubc_2a9,
im:7141966, tdsubc_2a9, wu:fb37c02, wu:fb73e06, xx:tdsubc_2a9,
zgc:56504; pre-mRNA processing factor 8; K12856 pre-mRNA-processing
factor 8
Length=2342
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T++DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 480 FRSFKATKFFQSTKLDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 539
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCRE+LR KL+ D
Sbjct 540 KERKKSRFGNAFHLCREVLRLSKLVVD 566
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 32/42 (76%), Gaps = 2/42 (4%)
Query 62 ERKKSRFGNA--FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
E++K F +A + E +RK+I+DHGDM++RK+RHD+RVY
Sbjct 54 EKRKFGFVDAQKEDMPPEHVRKIIRDHGDMTNRKFRHDKRVY 95
> hsa:10594 PRPF8, HPRP8, PRP8, PRPC8, RP13; PRP8 pre-mRNA processing
factor 8 homolog (S. cerevisiae); K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T++DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 473 FRSFKATKFFQSTKLDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 532
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCRE+LR KL+ D
Sbjct 533 KERKKSRFGNAFHLCREVLRLTKLVVD 559
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 32/42 (76%), Gaps = 2/42 (4%)
Query 62 ERKKSRFGNA--FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
E++K F +A + E +RK+I+DHGDM++RK+RHD+RVY
Sbjct 47 EKRKFGFVDAQKEDMPPEHVRKIIRDHGDMTNRKFRHDKRVY 88
> xla:379945 prpf8, MGC52804, hprp8, prp-8, prp8, prpc8, rp13;
PRP8 pre-mRNA processing factor 8 homolog; K12856 pre-mRNA-processing
factor 8
Length=2335
Score = 154 bits (390), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 79/87 (90%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR FK TKFFQ T++DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 473 FRSFKATKFFQSTKLDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 532
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCRE+LR KL+ D
Sbjct 533 KERKKSRFGNAFHLCREVLRLTKLVVD 559
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 32/42 (76%), Gaps = 2/42 (4%)
Query 62 ERKKSRFGNA--FHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
E++K F +A + E +RK+I+DHGDM++RK+RHD+RVY
Sbjct 47 EKRKFGFVDAQKEDMPPEHVRKIIRDHGDMTNRKFRHDKRVY 88
> ath:AT4G38780 splicing factor, putative
Length=2332
Score = 154 bits (388), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 77/87 (88%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR TKFFQ TE+DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 471 FRSLAATKFFQSTELDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 530
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 531 KERKKSRFGNAFHLCREILRLTKLVVD 557
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 2/49 (4%)
Query 53 RPVKTLTTKERKKSRFGNAFHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
R T+ T E +++ ++ K +DHGDMSS+K+R D+RVY
Sbjct 37 RNTPTVPTPEDAEAKLEKKARTWMQLNSK--RDHGDMSSKKHRLDKRVY 83
> ath:AT1G80070 SUS2; SUS2 (ABNORMAL SUSPENSOR 2); K12856 pre-mRNA-processing
factor 8
Length=2382
Score = 153 bits (387), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 74/87 (85%), Positives = 77/87 (88%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
FR TKFFQ TE+DWVE GLQV RQGYNMLNLLIHRKNLNYLHLDYNFNL+PVKTLTT
Sbjct 519 FRSLAATKFFQSTELDWVEVGLQVCRQGYNMLNLLIHRKNLNYLHLDYNFNLKPVKTLTT 578
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCREILR KL+ D
Sbjct 579 KERKKSRFGNAFHLCREILRLTKLVVD 605
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/20 (75%), Positives = 19/20 (95%), Gaps = 0/20 (0%)
Query 82 LIKDHGDMSSRKYRHDRRVY 101
L +DHGDMSS+K+RHD+RVY
Sbjct 116 LARDHGDMSSKKFRHDKRVY 135
> cpv:cgd3_2890 Prp8. JAB/PAD domain ; K12856 pre-mRNA-processing
factor 8
Length=2379
Score = 147 bits (371), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 68/87 (78%), Positives = 77/87 (88%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
++ + TKFFQ TE+DWVE GLQV +QGYNMLNLLIHRKNL YLHLDYNFNL+P+KTLTT
Sbjct 463 LKILQNTKFFQSTEMDWVEVGLQVCKQGYNMLNLLIHRKNLTYLHLDYNFNLKPIKTLTT 522
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSRFGNAFHLCRE+LR KL+ D
Sbjct 523 KERKKSRFGNAFHLCREVLRLTKLVVD 549
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/28 (64%), Positives = 23/28 (82%), Gaps = 0/28 (0%)
Query 74 LCREILRKLIKDHGDMSSRKYRHDRRVY 101
L E LRKLI HGDMSSRK+++++RVY
Sbjct 49 LPPEHLRKLINIHGDMSSRKFQNEKRVY 76
> sce:YHR165C PRP8, DBF3, DNA39, RNA8, SLT21, USA2; Component
of the U4/U6-U5 snRNP complex, involved in the second catalytic
step of splicing; mutations of human Prp8 cause retinitis
pigmentosa; K12856 pre-mRNA-processing factor 8
Length=2413
Score = 132 bits (332), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 64/87 (73%), Positives = 72/87 (82%), Gaps = 2/87 (2%)
Query 1 FRVFKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTT 60
+ K TK+FQ T IDWVEAGLQ+ RQG+NMLNLLIHRK L YLHLDYNFNL+P KTLTT
Sbjct 548 LKSLKNTKYFQQTTIDWVEAGLQLCRQGHNMLNLLIHRKGLTYLHLDYNFNLKPTKTLTT 607
Query 61 KERKKSRFGNAFHLCREILR--KLIKD 85
KERKKSR GN+FHL RE+L+ KLI D
Sbjct 608 KERKKSRLGNSFHLMRELLKMMKLIVD 634
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 55 VKTLTTKERKKSRFGNAFHLCREILRKLIKDHGDMSSRKYRHDRRVY 101
K +T K ++ + + + E LRK+I H DM+S+ Y D++ +
Sbjct 115 AKKMTKKAKRSNLYTPKAEMPPEHLRKIINTHSDMASKMYNTDKKAF 161
> mmu:113851 Vmn1r54, V1ra9, VN7; vomeronasal 1 receptor 54; K04614
vomeronasal1 receptor
Length=315
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query 4 FKTTKFFQLTEIDWVEAGLQVSRQGYNMLNLLIHRKNLNYLHLDYNFNLRP 54
F T F LT D + GL + GY + L H+K +LH+ +F+L+P
Sbjct 181 FHLTVFILLTSRDVIFVGLMLLSSGYMVTFLGRHKKQSQFLHIT-SFSLKP 230
> mmu:100043597 Srcap, B930091H02Rik, D030022P06Rik, F630004O05Rik;
Snf2-related CREBBP activator protein; K11661 helicase
SRCAP [EC:3.6.4.-]
Length=3237
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 4/40 (10%)
Query 61 KERKKSRFG----NAFHLCREILRKLIKDHGDMSSRKYRH 96
KERK R G NAFH+C + +++DH + +R+
Sbjct 715 KERKLKRQGWTKPNAFHVCITSYKLVLQDHQAFRRKNWRY 754
> hsa:10847 SRCAP, DOMO1, EAF1, FLJ44499, KIAA0309, SWR1; Snf2-related
CREBBP activator protein; K11661 helicase SRCAP [EC:3.6.4.-]
Length=3230
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 4/40 (10%)
Query 61 KERKKSRFG----NAFHLCREILRKLIKDHGDMSSRKYRH 96
KERK R G NAFH+C + +++DH + +R+
Sbjct 703 KERKLKRQGWTKPNAFHVCITSYKLVLQDHQAFRRKNWRY 742
> hsa:83889 WDR87, FLJ52608, NYD-SP11; WD repeat domain 87
Length=2873
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 11/46 (23%)
Query 12 LTEIDWV-------EAGLQVSRQGYN----MLNLLIHRKNLNYLHL 46
L++++W+ EAG Q+SR G++ +L L + NL +LHL
Sbjct 2522 LSDVEWIRHVLERMEAGEQLSRDGFHRLCQLLKDLASKGNLEWLHL 2567
> cpv:cgd5_820 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=523
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query 33 NLLIHRKNLNYLHLDYNFNLRPVKTLTTKERKKSRFGNAFHLCREILRKLIKDHGDMSS 91
NLL+ K + L +F L V +++ K R G A+++ E+LRK + D+ S
Sbjct 189 NLLLESKEKDALIKIVDFGLSAV--FENQKKMKERLGTAYYIAPEVLRKKYDEKCDVWS 245
> dre:100151066 MGC195210; cardiomyopathy associated 5 like
Length=1759
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 11/75 (14%)
Query 27 QGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTTKERKKSRF----GNAFHLCREILRKL 82
+GY LL + N NYL F ++ V + E+ ++ G FH RE +
Sbjct 1540 KGYEQRVLL--QPNENYL-----FYIKSVNAGGSSEQSEAALISTRGTRFHFLRESAHSV 1592
Query 83 IKDHGDMSSRKYRHD 97
+K D +S +Y HD
Sbjct 1593 LKVSEDRNSVEYPHD 1607
> dre:560687 cmya5, cmya5l; cardiomyopathy associated 5
Length=1759
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 11/75 (14%)
Query 27 QGYNMLNLLIHRKNLNYLHLDYNFNLRPVKTLTTKERKKSRF----GNAFHLCREILRKL 82
+GY LL + N NYL F ++ V + E+ ++ G FH RE +
Sbjct 1540 KGYEQRVLL--QPNENYL-----FYIKSVNAGGSSEQSEAALISTRGTRFHFLRESAHSV 1592
Query 83 IKDHGDMSSRKYRHD 97
+K D +S +Y HD
Sbjct 1593 LKVSEDRNSVEYPHD 1607
Lambda K H
0.328 0.141 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40